

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.4 - phase: 0
(338 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
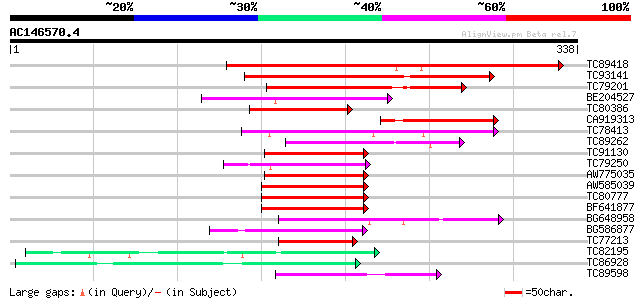
Score E
Sequences producing significant alignments: (bits) Value
TC89418 weakly similar to GP|10800070|dbj|BAB16490. contains EST... 164 4e-41
TC93141 similar to GP|18491261|gb|AAL69455.1 At2g41130/T3K9.10 {... 164 6e-41
TC79201 weakly similar to GP|10800070|dbj|BAB16490. contains EST... 107 9e-24
BE204527 similar to GP|10800070|dbj contains ESTs C61063(AU06336... 80 1e-15
TC80386 weakly similar to GP|10800070|dbj|BAB16490. contains EST... 70 9e-13
CA919313 62 2e-10
TC78413 similar to PIR|T06329|T06329 symbiotic ammonium transpor... 62 4e-10
TC89262 similar to GP|21537346|gb|AAM61687.1 unknown {Arabidopsi... 58 6e-09
TC91130 homologue to GP|11245494|gb|AAG33640.1 SPATULA {Arabidop... 57 8e-09
TC79250 similar to PIR|T06329|T06329 symbiotic ammonium transpor... 55 4e-08
AW775035 similar to GP|13486760|dbj hypothetical protein {Oryza ... 55 5e-08
AW585039 similar to GP|13605859|gb AT4g02590/T10P11_13 {Arabidop... 54 7e-08
TC80777 homologue to GP|4115386|gb|AAD03387.1| unknown protein {... 54 7e-08
BF641877 similar to GP|13605859|gb AT4g02590/T10P11_13 {Arabidop... 54 1e-07
BG648958 similar to GP|19310467|gb| At1g01260/F6F3_25 {Arabidops... 54 1e-07
BG586877 similar to GP|16226919|gb AT4g16430/dl4240w {Arabidopsi... 53 1e-07
TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding pro... 53 2e-07
TC82195 homologue to GP|19310475|gb|AAL84972.1 AT3g26744/MLJ15_1... 52 4e-07
TC86928 similar to PIR|E96714|E96714 probable DNA-binding protei... 51 6e-07
TC89598 weakly similar to GP|20127014|gb|AAM10934.1 putative bHL... 51 7e-07
>TC89418 weakly similar to GP|10800070|dbj|BAB16490. contains ESTs
C61063(AU063369) C61063(AU093310)~unknown protein,
partial (26%)
Length = 1093
Score = 164 bits (415), Expect = 4e-41
Identities = 87/207 (42%), Positives = 132/207 (63%), Gaps = 6/207 (2%)
Frame = +3
Query: 130 SGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTK 189
S SG F + + + + +E +A +ASKSHS+AE+RRR+RIN LA LR L+P + K
Sbjct: 132 SSSGSFQYSEFQSWNLPIEGSVEDRAASASKSHSQAEKRRRDRINTQLANLRKLIPKSDK 311
Query: 190 TDKASLLAEVIQHVKELKRQTSLIAETSPVPTECDELTVD--AAADDEDYGSNGNKF--- 244
DKA+LL VI VK+LKR+ ++ VPTE DE+++D +DE + NKF
Sbjct: 312 MDKAALLGSVIDQVKDLKRKAMDVSRVITVPTEIDEVSIDYNHVVEDETNTNKVNKFKDN 491
Query: 245 -IIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLFITGEEDDHEYCIS 303
IIKAS+CCDDR +L ELI+ LK+LRL T+KADI ++GGR+K++L + ++ + CI+
Sbjct: 492 IIIKASVCCDDRPELFSELIQVLKSLRLTTVKADIASVGGRIKSILVLCSKDSEENVCIN 671
Query: 304 SIQEALKAVMEKSVGDESASGSVKRQR 330
+++++LK+ + K S R +
Sbjct: 672 TLKQSLKSAVTKIASSSMVSNCPTRSK 752
>TC93141 similar to GP|18491261|gb|AAL69455.1 At2g41130/T3K9.10 {Arabidopsis
thaliana}, partial (57%)
Length = 839
Score = 164 bits (414), Expect = 6e-41
Identities = 85/149 (57%), Positives = 113/149 (75%)
Frame = +2
Query: 141 EVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVI 200
EV +++ + +ALAA K+H EAE+RRRERIN+HL LR+LLP +KTDKASLLA+V+
Sbjct: 344 EVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTDKASLLAKVV 523
Query: 201 QHVKELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLP 260
+ VKELK+QTS I + VP+E DE+TV +A D S + I KASLCC+DRSDL+P
Sbjct: 524 ERVKELKQQTSQITQLETVPSETDEITVISAGSDI---SGEGRLIFKASLCCEDRSDLIP 694
Query: 261 ELIKTLKALRLRTLKADITTLGGRVKNVL 289
+LI+ LK+L L+TLKA++ TLGGR +NVL
Sbjct: 695 DLIEILKSLHLKTLKAEMATLGGRTRNVL 781
>TC79201 weakly similar to GP|10800070|dbj|BAB16490. contains ESTs
C61063(AU063369) C61063(AU093310)~unknown protein,
partial (18%)
Length = 951
Score = 107 bits (266), Expect = 9e-24
Identities = 60/119 (50%), Positives = 80/119 (66%)
Frame = +3
Query: 154 KALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLI 213
K L A K+H EAER+RR RIN HLAKLR+L+PS+ K DKA+LLAEVI+ VK LK+
Sbjct: 258 KTLVALKNHREAERKRRNRINGHLAKLRALVPSSPKMDKATLLAEVIRQVKHLKKNADEA 437
Query: 214 AETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLR 272
++ +PT+ DE+ V E Y NG F+ KAS+ CD R +LL +L +TL L+L+
Sbjct: 438 SKGYSIPTDDDEVKV------EPY-ENGGSFLYKASISCDYRPELLSDLRQTLDKLQLQ 593
>BE204527 similar to GP|10800070|dbj contains ESTs C61063(AU063369)
C61063(AU093310)~unknown protein, partial (18%)
Length = 579
Score = 79.7 bits (195), Expect = 1e-15
Identities = 48/122 (39%), Positives = 69/122 (56%), Gaps = 8/122 (6%)
Frame = +3
Query: 115 GSEHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEI-MEAKALA-------ASKSHSEAE 166
G + ++ + G GS E G++ + + ME K ++ A K+HSEAE
Sbjct: 213 GKNGFEVNNEVIGSGYGSSFSLVLDRERGELVEEPVKMEGKGVSTTERSVEALKNHSEAE 392
Query: 167 RRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECDEL 226
RRRR RIN HL LR ++P K DKASLL EV++H+KELKR + E +P + DE+
Sbjct: 393 RRRRARINAHLDTLRCVIPGALKMDKASLLGEVVRHLKELKRNETQACEGLMIPKDNDEI 572
Query: 227 TV 228
+V
Sbjct: 573 SV 578
>TC80386 weakly similar to GP|10800070|dbj|BAB16490. contains ESTs
C61063(AU063369) C61063(AU093310)~unknown protein,
partial (17%)
Length = 655
Score = 70.5 bits (171), Expect = 9e-13
Identities = 34/61 (55%), Positives = 45/61 (73%)
Frame = +1
Query: 144 KMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHV 203
K+ + + +++ A K+HSEAERRRR RIN+HL LRS++P K DKASLLAEVI H+
Sbjct: 472 KLERKGVSPERSIEALKNHSEAERRRRARINSHLDTLRSVIPGANKLDKASLLAEVITHL 651
Query: 204 K 204
K
Sbjct: 652 K 654
>CA919313
Length = 786
Score = 62.4 bits (150), Expect = 2e-10
Identities = 32/70 (45%), Positives = 46/70 (65%)
Frame = -1
Query: 222 ECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTL 281
+CDE+ V++ ++ G NG + I+ASLCC + DLL ++ K L AL +KA I TL
Sbjct: 447 DCDEIRVES----QEGGLNGFPYSIRASLCCQYKPDLLSDIRKALDALHPMIIKAKIATL 280
Query: 282 GGRVKNVLFI 291
GGR+KNV+ I
Sbjct: 279 GGRIKNVVVI 250
>TC78413 similar to PIR|T06329|T06329 symbiotic ammonium transport protein
SAT1 - soybean, partial (30%)
Length = 1397
Score = 61.6 bits (148), Expect = 4e-10
Identities = 43/181 (23%), Positives = 87/181 (47%), Gaps = 28/181 (15%)
Frame = +2
Query: 139 QGEVGKMSAQEIMEA---------KALAASKSHSEAERRRRERINNHLAKLRSLLPSTTK 189
Q G + Q I+E +++A ++ H AER+RRE+++ L L +L+P K
Sbjct: 437 QNRKGSLQKQNIVETIKPQGQGTKRSVAHNQDHIIAERKRREKLSQCLIALAALIPGLKK 616
Query: 190 TDKASLLAEVIQHVKELKRQTSLIAE-------TSPVPTECDELTVDAA-ADDEDYGSNG 241
DKAS+L + I++VKEL+ + ++ E S V + +L+ D++ +DD + S
Sbjct: 617 MDKASVLGDAIKYVKELQERLRVLEEQNKNSHVQSVVTVDEQQLSYDSSNSDDSEVASGN 796
Query: 242 NKFI-----------IKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLF 290
N+ + + + C + LL +++ ++ L L + + G + ++
Sbjct: 797 NETLPHVEAKVLDKDVLIRIHCQKQKGLLLKILVEIQKLHLFVVNNSVLPFGDSILDITI 976
Query: 291 I 291
+
Sbjct: 977 V 979
>TC89262 similar to GP|21537346|gb|AAM61687.1 unknown {Arabidopsis
thaliana}, partial (40%)
Length = 1198
Score = 57.8 bits (138), Expect = 6e-09
Identities = 34/113 (30%), Positives = 61/113 (53%), Gaps = 6/113 (5%)
Frame = +3
Query: 165 AERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECD 224
AERRRR+R+N+ L+ LR+++P +K D+ ++L + I ++KEL + + + + +
Sbjct: 567 AERRRRKRLNDRLSMLRAIVPKISKMDRTAILGDTIDYMKELLEKIKNLQQEIELDSNMT 746
Query: 225 ELTVDAAADDEDYGSNGNKFIIKAS------LCCDDRSDLLPELIKTLKALRL 271
+ D +E N KF ++ S +CC + LL + TL+AL L
Sbjct: 747 SIVKD-VKPNEILIRNSPKFEVERSADTRVEICCAGKPGLLLSTVNTLEALGL 902
>TC91130 homologue to GP|11245494|gb|AAG33640.1 SPATULA {Arabidopsis
thaliana}, partial (20%)
Length = 766
Score = 57.4 bits (137), Expect = 8e-09
Identities = 25/62 (40%), Positives = 46/62 (73%)
Frame = +2
Query: 153 AKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSL 212
+K A++ H+ +E+RRR RIN + L++L+P++ KTDKAS+L E I+++K+L+ Q +
Sbjct: 17 SKRSRAAEVHNLSEKRRRSRINEKMKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQM 196
Query: 213 IA 214
++
Sbjct: 197 LS 202
>TC79250 similar to PIR|T06329|T06329 symbiotic ammonium transport protein
SAT1 - soybean, partial (34%)
Length = 1145
Score = 55.1 bits (131), Expect = 4e-08
Identities = 33/92 (35%), Positives = 49/92 (52%), Gaps = 4/92 (4%)
Frame = +2
Query: 128 HGSGSGPFGGFQGEVGKMSAQEIMEAK----ALAASKSHSEAERRRRERINNHLAKLRSL 183
HG G + + K + I E+K A ++ H AER+RRE+I+ L +L
Sbjct: 401 HGKSLASKGSLENQK-KGPKRNIQESKKTDSAARNAQDHIIAERKRREKISQKFIALSAL 577
Query: 184 LPSTTKTDKASLLAEVIQHVKELKRQTSLIAE 215
LP K DKAS+L + I HVK+L+ + L+ E
Sbjct: 578 LPDLKKMDKASVLGDAINHVKQLQEKVKLLEE 673
>AW775035 similar to GP|13486760|dbj hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 716
Score = 54.7 bits (130), Expect = 5e-08
Identities = 25/62 (40%), Positives = 43/62 (69%)
Frame = +1
Query: 153 AKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSL 212
+K A++ H+ +ERRRR+RIN + L+ L+P+ K DKAS+L E I+++K L+ Q +
Sbjct: 481 SKRSRAAEVHNLSERRRRDRINEKMRALQELIPNCNKVDKASMLDEAIEYLKTLQLQVQM 660
Query: 213 IA 214
++
Sbjct: 661 MS 666
>AW585039 similar to GP|13605859|gb AT4g02590/T10P11_13 {Arabidopsis
thaliana}, partial (46%)
Length = 628
Score = 54.3 bits (129), Expect = 7e-08
Identities = 27/64 (42%), Positives = 42/64 (65%)
Frame = +1
Query: 151 MEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQT 210
+ A+ A+ HS AER RRERI + L+ L+PS KTDKA++L E++ +VK L+ Q
Sbjct: 151 VRARRGQATDPHSIAERLRRERIAERMKALQELVPSINKTDKAAMLDEIVDYVKFLRLQV 330
Query: 211 SLIA 214
+++
Sbjct: 331 KVLS 342
>TC80777 homologue to GP|4115386|gb|AAD03387.1| unknown protein {Arabidopsis
thaliana}, partial (39%)
Length = 1307
Score = 54.3 bits (129), Expect = 7e-08
Identities = 28/64 (43%), Positives = 42/64 (64%)
Frame = +1
Query: 151 MEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQT 210
+ A+ A+ HS AER RRERI + L+ L+P+ KTDKAS+L E+I +VK L+ Q
Sbjct: 436 VRARRGQATDPHSIAERLRRERIAERMKALQELVPNANKTDKASMLDEIIDYVKFLQVQV 615
Query: 211 SLIA 214
+++
Sbjct: 616 KVLS 627
>BF641877 similar to GP|13605859|gb AT4g02590/T10P11_13 {Arabidopsis
thaliana}, partial (55%)
Length = 642
Score = 53.5 bits (127), Expect = 1e-07
Identities = 26/64 (40%), Positives = 41/64 (63%)
Frame = +2
Query: 151 MEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQT 210
+ A+ A+ HS AER RRERI + L+ L+PS KTD+A++L E++ +VK L+ Q
Sbjct: 449 VRARRGQATDPHSIAERLRRERIAERIRALQELVPSVNKTDRAAMLXEIVDYVKFLRLQV 628
Query: 211 SLIA 214
++
Sbjct: 629 KXLS 640
>BG648958 similar to GP|19310467|gb| At1g01260/F6F3_25 {Arabidopsis
thaliana}, partial (15%)
Length = 806
Score = 53.5 bits (127), Expect = 1e-07
Identities = 37/147 (25%), Positives = 72/147 (48%), Gaps = 13/147 (8%)
Frame = +3
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLI---AETS 217
+H EAER+RRE++N LR+++P+ +K DKASLL + I ++ EL+ + ++ E+
Sbjct: 330 NHVEAERQRREKLNQRFYALRAVVPNISKMDKASLLGDAIAYINELQAKLKIMESERESF 509
Query: 218 PVPTECDELTVDAAAD----------DEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLK 267
T D L + + D D ++ ++ I+K S D + ++I+T K
Sbjct: 510 GSSTSRDGLANSRSENRHQNTRVPPPDVDIQASQDEVIVKVSCSLDTHP--VSKVIETFK 683
Query: 268 ALRLRTLKADITTLGGRVKNVLFITGE 294
++ +++ + + I E
Sbjct: 684 EAQISVVESKLAAANDTIYPYFVIKSE 764
>BG586877 similar to GP|16226919|gb AT4g16430/dl4240w {Arabidopsis thaliana},
partial (35%)
Length = 755
Score = 53.1 bits (126), Expect = 1e-07
Identities = 32/94 (34%), Positives = 50/94 (53%)
Frame = +1
Query: 120 RIISDTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAK 179
R+ S L + S P G E K + A +H EAER+RRE++N
Sbjct: 484 RVSSIDLGNEDSSSPLG----EERKPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYA 651
Query: 180 LRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLI 213
LR+ +P+ +K DKASLL + I H+ +L+++ L+
Sbjct: 652 LRAGVPNISKMDKASLLGDAITHITDLQKKIKLM 753
>TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding protein PG1 -
kidney bean, partial (70%)
Length = 2391
Score = 52.8 bits (125), Expect = 2e-07
Identities = 23/47 (48%), Positives = 35/47 (73%)
Frame = +3
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELK 207
+H EAER+RRE++N LR+++P+ +K DKASLL + I ++ ELK
Sbjct: 1545 NHVEAERQRREKLNQKFYALRAVVPNVSKMDKASLLGDAISYINELK 1685
>TC82195 homologue to GP|19310475|gb|AAL84972.1 AT3g26744/MLJ15_15
{Arabidopsis thaliana}, partial (17%)
Length = 1189
Score = 51.6 bits (122), Expect = 4e-07
Identities = 64/243 (26%), Positives = 98/243 (39%), Gaps = 32/243 (13%)
Frame = +2
Query: 10 SSQTINNFETYQQEQFLLQQQMMRQQNSDHHIFGGGR---SMFPTAGGDHNQVSPILQQT 66
++ T N FE + F M S +F GG S FP + P +
Sbjct: 461 NNNTNNPFEMGFENGFF----MGNNNTSTSPVFMGGSLSASEFPPSLELDAAPVPPFSAS 628
Query: 67 WSMQ------QLHHHHHPFNPHDPFVIPQQQASPYASLFNRRVPSLQFAYDHHAGSEHLR 120
+SM Q HHH QQQ +LF +R +L+ G++ R
Sbjct: 629 FSMPLELAQPQPQQHHH-----------QQQQQQPTTLFQKRRGALEIPRLETVGNKKKR 775
Query: 121 IISDTLQHGSGSGPFGG----------------------FQGEVGKMSAQEIMEAKALAA 158
+ + + GSG GG G V Q+ + K L A
Sbjct: 776 KVEKSWEE-EGSGGGGGDDVDDFSELNYDSDENGNDLNNSNGTVVTGGDQKGKKKKGLPA 952
Query: 159 SKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKE-LKRQTSLIAETS 217
+ AERRRR+++N+ L LRS++P +K D+AS+L + + ++KE L+R +L E
Sbjct: 953 K--NLMAERRRRKKLNDRLYMLRSVVPKISKMDRASILGDAVDYLKELLQRINNLHNELE 1126
Query: 218 PVP 220
P
Sbjct: 1127STP 1135
>TC86928 similar to PIR|E96714|E96714 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (39%)
Length = 1926
Score = 51.2 bits (121), Expect = 6e-07
Identities = 47/207 (22%), Positives = 79/207 (37%), Gaps = 1/207 (0%)
Frame = +3
Query: 4 EDQGQCSSQTINNFETYQQEQFLLQQQMMRQQNSDHHIFGGGRSMFPTAGGDHNQVSPIL 63
+D CSS T + F +++ + + D I G + L
Sbjct: 165 DDNSNCSSHTSSGFTLWEESEIRKGKTGKENNVGDKPIIGDSAAKLGQW---------TL 317
Query: 64 QQTWSMQQLHHHHHPFNPHDPFVIPQQQASPYASLFNRRVPSLQFAYDHHAGSEHLRIIS 123
++ S ++HH FN Q+ + + Q + G+ L+ S
Sbjct: 318 KERTSQSFSNNHHGSFNSRSSSQTTGQKNQSFMEMMKSAQDCAQDEELENEGTFFLKRES 497
Query: 124 DTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSL 183
+Q +GE+ + + K HS E+RRR +IN+ LR L
Sbjct: 498 SNVQ-----------RGELRVRVDGKSTDQKPNTPRSKHSATEQRRRSKINDRFQMLREL 644
Query: 184 LP-STTKTDKASLLAEVIQHVKELKRQ 209
+P S K DKAS L EVI+++ L+ +
Sbjct: 645 IPHSDQKRDKASFLLEVIEYIHFLQEK 725
>TC89598 weakly similar to GP|20127014|gb|AAM10934.1 putative bHLH
transcription factor {Arabidopsis thaliana}, partial
(24%)
Length = 849
Score = 50.8 bits (120), Expect = 7e-07
Identities = 28/99 (28%), Positives = 48/99 (48%)
Frame = +3
Query: 159 SKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSP 218
++ H AER+RRE+++ L S+LP K DKA++L + I+H+K+L+ + +
Sbjct: 531 ARDHVIAERKRREKLSQRFIALSSILPGLKKMDKATILEDAIKHMKQLQERVKTL----- 695
Query: 219 VPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSD 257
+E D + + F S CD+ SD
Sbjct: 696 -----EEQVADKKVESAVFVKRSILFDNNDSSSCDENSD 797
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,356,674
Number of Sequences: 36976
Number of extensions: 161864
Number of successful extensions: 2464
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 2077
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2337
length of query: 338
length of database: 9,014,727
effective HSP length: 97
effective length of query: 241
effective length of database: 5,428,055
effective search space: 1308161255
effective search space used: 1308161255
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146570.4


