

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
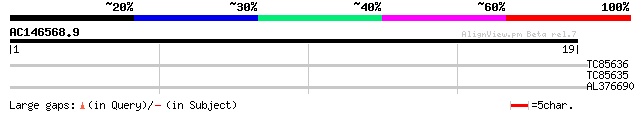
Query= AC146568.9 + phase: 0 /pseudo
(19 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85636 homologue to GP|4959710|gb|AAD34458.1| Skp1 {Medicago sa... 36 0.003
TC85635 homologue to GP|4959710|gb|AAD34458.1| Skp1 {Medicago sa... 36 0.003
AL376690 similar to PIR|T02710|T027 putative kinetechore (Skp1p-... 31 0.13
>TC85636 homologue to GP|4959710|gb|AAD34458.1| Skp1 {Medicago sativa},
complete
Length = 898
Score = 36.2 bits (82), Expect = 0.003
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = +1
Query: 3 SSTRKITLKSSDGETFE 19
SSTRKITLKSSDGETFE
Sbjct: 139 SSTRKITLKSSDGETFE 189
>TC85635 homologue to GP|4959710|gb|AAD34458.1| Skp1 {Medicago sativa},
complete
Length = 914
Score = 36.2 bits (82), Expect = 0.003
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = +3
Query: 3 SSTRKITLKSSDGETFE 19
SSTRKITLKSSDGETFE
Sbjct: 135 SSTRKITLKSSDGETFE 185
>AL376690 similar to PIR|T02710|T027 putative kinetechore (Skp1p-like)
protein [imported] - Arabidopsis thaliana, partial (22%)
Length = 402
Score = 30.8 bits (68), Expect = 0.13
Identities = 14/19 (73%), Positives = 16/19 (83%)
Frame = +3
Query: 1 MSSSTRKITLKSSDGETFE 19
+ SST+KITL SSDGE FE
Sbjct: 111 VESSTKKITLTSSDGEIFE 167
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.298 0.112 0.266
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 389,512
Number of Sequences: 36976
Number of extensions: 925
Number of successful extensions: 4
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4
length of query: 19
length of database: 9,014,727
effective HSP length: 0
effective length of query: 24
effective length of database: 9,014,727
effective search space: 216353448
effective search space used: 216353448
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146568.9


