

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.7 - phase: 0
(703 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
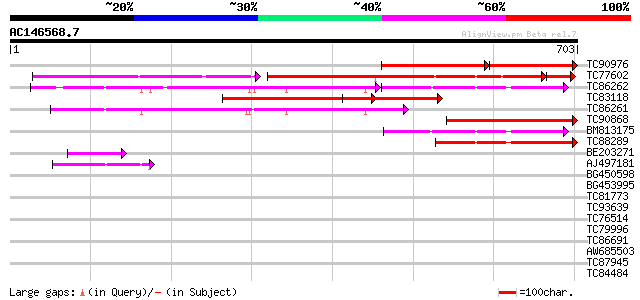
Score E
Sequences producing significant alignments: (bits) Value
TC90976 similar to PIR|E96718|E96718 probable cullin T6C23.13 [i... 276 e-131
TC77602 similar to GP|20268719|gb|AAM14063.1 putative cullin {Ar... 307 e-122
TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 220 2e-97
TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [i... 291 6e-79
TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 273 2e-73
TC90868 similar to GP|9295728|gb|AAF87034.1| T24P13.25 {Arabidop... 251 9e-67
BM813175 similar to GP|22136936|gb putative cullin 1 protein {Ar... 157 2e-38
TC88289 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 129 3e-30
BE203271 similar to GP|22550314|gb| putative cullin protein {Ole... 49 9e-06
AJ497181 weakly similar to GP|22335691|dbj cullin-like protein1 ... 43 5e-04
BG450598 similar to GP|23429518|gb APC2 {Arabidopsis thaliana}, ... 40 0.004
BG453995 weakly similar to GP|14091836|gb| Putative cullin {Oryz... 40 0.004
TC81773 homologue to PIR|E96718|E96718 probable cullin T6C23.13 ... 38 0.012
TC93639 similar to GP|22335691|dbj|BAC10548. cullin-like protein... 36 0.044
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 34 0.17
TC79996 similar to PIR|T00571|T00571 dolichyl-phosphate beta-glu... 33 0.49
TC86691 similar to GP|15810175|gb|AAL06989.1 At2g03890/T18C20.9 ... 32 1.1
AW685503 similar to GP|22165062|gb| unknown protein {Oryza sativ... 32 1.1
TC87945 weakly similar to GP|17473713|gb|AAL38309.1 unknown prot... 31 1.4
TC84484 similar to GP|20161418|dbj|BAB90342. phospholipase-like ... 30 3.2
>TC90976 similar to PIR|E96718|E96718 probable cullin T6C23.13 [imported] -
Arabidopsis thaliana, partial (33%)
Length = 825
Score = 276 bits (706), Expect(2) = e-131
Identities = 135/135 (100%), Positives = 135/135 (100%)
Frame = +3
Query: 461 TLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKAT 520
TLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKAT
Sbjct: 3 TLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKAT 182
Query: 521 FGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVL 580
FGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVL
Sbjct: 183 FGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVL 362
Query: 581 RKEPMSKDVGEDDAF 595
RKEPMSKDVGEDDAF
Sbjct: 363 RKEPMSKDVGEDDAF 407
Score = 212 bits (540), Expect(2) = e-131
Identities = 109/109 (100%), Positives = 109/109 (100%)
Frame = +1
Query: 595 FSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDH 654
FSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDH
Sbjct: 406 FSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDH 585
Query: 655 NNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
NNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA
Sbjct: 586 NNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 732
>TC77602 similar to GP|20268719|gb|AAM14063.1 putative cullin {Arabidopsis
thaliana}, partial (87%)
Length = 2708
Score = 307 bits (786), Expect(2) = e-122
Identities = 167/350 (47%), Positives = 241/350 (68%), Gaps = 4/350 (1%)
Frame = +2
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSP-EFISLFVDDKLRKGLKGVNEDDVEVTLDKVM 378
++F +++F N + +FE+ INL P E I+ F+DDKLR G KG +E+++E TLDKV+
Sbjct: 968 ESFAKNEAFSNTIKDAFEHLINLRQNRPAELIAKFLDDKLRAGNKGTSEEELEGTLDKVL 1147
Query: 379 MLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTD 438
+LFR++Q KDVFE +YK+ LAKRLL GK+ S DAE+S+I KLKTECG QFT+KLEGMF D
Sbjct: 1148 VLFRFIQGKDVFEAFYKKDLAKRLLLGKSASIDAEKSMISKLKTECGSQFTNKLEGMFKD 1327
Query: 439 MKTSQDTMQGFYAS---HPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRS 495
++ S++ + F S L G ++V VLTTG WPT + LP E++ + F+
Sbjct: 1328 IELSKEINESFRQSSQARTKLPSGIEMSVHVLTTGYWPTYPPMDVRLPHELNVYQDIFKE 1507
Query: 496 YYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQ 555
+YL ++GRRL WQ ++G LKA F KG+K EL VS +Q VLM FN+A+KLS+++I+
Sbjct: 1508 FYLSKYSGRRLMWQNSLGHCVLKADFPKGKK-ELAVSLFQTVVLMQFNDAEKLSFQDIKD 1684
Query: 556 ATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVA 615
+T I +L+R LQSLA K R VL+K P +DV + D+F ND F++ LY++K+ +
Sbjct: 1685 STGIEDKELRRTLQSLACGKVR-VLQKMPKGRDVEDYDSFVFNDTFTAPLYRIKV-NAIQ 1858
Query: 616 QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQL 665
KE+ E T +RV +DR+ Q++AAIVRIMK+R++L H LI E+ +Q+
Sbjct: 1859 LKETVEENTNTTERVFQDRQYQVDAAIVRIMKTRKVLSHTLLITELFQQV 2008
Score = 151 bits (382), Expect(2) = e-122
Identities = 89/286 (31%), Positives = 158/286 (55%), Gaps = 3/286 (1%)
Frame = +3
Query: 29 NMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGS--FLEELNRKWNDHNKALQMIRD 86
++ +HK G LY + H+ +S+ FL + R W D + MIR
Sbjct: 111 DLCIHKMGGNLYQRIEKECEVHISAALQSLVGQSPDLIVFLSLVERCWQDLCDQMLMIRG 290
Query: 87 ILMYMDRTFIPSAKKT-PVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDR 145
I +++DRT++ + + ++GL ++R+ + S +++ + + LL ++ SER GE +DR
Sbjct: 291 IALFLDRTYVKQSPNIRSIWDMGLQIFRKHLSLSPEVQHKTVTGLLRMIDSERLGEAVDR 470
Query: 146 GIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNE 205
++ ++ KM LG +Y + FE FL+ ++EFY E ++++ D DYLK E RL E
Sbjct: 471 TLLNHLLKMFTALG--IYAESFEKPFLECTSEFYAAEGVKYMQQSDVPDYLKHVETRLQE 644
Query: 206 EMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLF 265
E +R Y+D T+K + E Q++E H+ ++ + G ++ ++ EDL RM+ LF
Sbjct: 645 EHERCLIYLDASTKKPLITTTEKQLLERHIPAIL---DKGFSMLMDGNRIEDLQRMHLLF 815
Query: 266 RRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEK 311
RV + L +R+ ++ +IR +G+ +V D E+ KD VQ LL+ K
Sbjct: 816 SRV-NALESLRQAISSYIRRTGQGIVMDEEKDKD---MVQSLLEFK 941
Score = 45.4 bits (106), Expect = 7e-05
Identities = 19/36 (52%), Positives = 29/36 (79%)
Frame = +3
Query: 666 QLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRY 701
QL+F P ++KKRIESLI+R++LERD ++ ++Y Y
Sbjct: 2346 QLKFPIKPADLKKRIESLIDREYLERDKSNPQVYNY 2453
>TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, complete
Length = 2332
Score = 220 bits (561), Expect(2) = 2e-97
Identities = 139/463 (30%), Positives = 246/463 (53%), Gaps = 29/463 (6%)
Frame = +1
Query: 26 NAYNMVLH-KFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMI 84
N Y+ L+ K+ + +V+T+ L+E L EL ++W +H ++ +
Sbjct: 289 NDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEF-------MLRELVKRWANHKIMVRWL 447
Query: 85 RDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVID 144
Y+DR FI P++E+GL +R+ V ++ ++ + ++ L+ ER GE ID
Sbjct: 448 SRFFHYLDRYFIARRSLPPLNEVGLACFRDLVY--KELHGKMRDAIISLIDQEREGEQID 621
Query: 145 RGIMRNITKMLMDLGPAV---YGQDFEAHFLQ-----VSAEFYQVESQRFIECCDCGDYL 196
R +++N+ + +++G Y DFEA L+ + ++ ++++ +RF+ Y
Sbjct: 622 RALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTICLLFSKGFKLDPRRFLS*L----YA 789
Query: 197 KKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYE 256
+ L E DRV HY+ +E K+ V+ +++ + +L+ E+SG +L DDK E
Sbjct: 790 SRLRSALRREKDRVAHYLHSSSEPKLLTKVQNELLSVYASQLLEKEHSGCHALLKDDKCE 969
Query: 257 DLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPE--------RLKDPVE-----F 303
DL RM+ LF ++ GL + + H+ G LV E + +D V F
Sbjct: 970 DLSRMFRLFSKIPSGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAQKRDIVGTHEQVF 1149
Query: 304 VQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFIN---LNPRSPEFISLFVDDKLRK 360
V+ +++ DKY +N N F AL +FE F N S E ++ F D+ L+K
Sbjct: 1150VRIVIELHDKYLAYVNSCLQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKK 1329
Query: 361 G-LKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVK 419
G + ++++ +E TL+KV+ L Y+ +KD+F ++Y++ LA+RLL K+ +DD ERS++ K
Sbjct: 1330GGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTK 1509
Query: 420 LKTECGYQFTSKLEGMFTDM---KTSQDTMQGFYASHPDLGDG 459
LK +CG QFTSK+EGM TD+ K +Q + + + ++ P+ G
Sbjct: 1510LKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPG 1638
Score = 154 bits (389), Expect(2) = 2e-97
Identities = 92/232 (39%), Positives = 135/232 (57%)
Frame = +2
Query: 461 TLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKAT 520
TLTV VLTTG WP+ S NLP E+ E F+ +Y R+L+W ++G ++
Sbjct: 1643 TLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGK 1822
Query: 521 FGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVL 580
F + EL V+TYQ L+LFN++D+LSY EI + D+ R L SL+ K + +L
Sbjct: 1823 FDP-KTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYK-IL 1996
Query: 581 RKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEA 640
KEP +K + D F N KF+ K+ ++KI EK++ + V++DR+ I+A
Sbjct: 1997 IKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDA 2161
Query: 641 AIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERD 692
+IVRIMKSR++L + L+ E + L F + +KKRIE LI RD+LERD
Sbjct: 2162 SIVRIMKSRKVLGYQQLVMECVEXLGRMFKPDVKAIKKRIEXLISRDYLERD 2317
>TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 902
Score = 291 bits (745), Expect = 6e-79
Identities = 156/192 (81%), Positives = 159/192 (82%), Gaps = 2/192 (1%)
Frame = +2
Query: 265 FRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNN 324
FRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNN
Sbjct: 2 FRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNN 181
Query: 325 DKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYL 384
DKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYL
Sbjct: 182 DKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYL 361
Query: 385 QEKDVFEKYYKQHLAKR--LLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTS 442
QEKDVFEKYYKQHLAK +L + + ERS FTSKLEGMFTDMKT
Sbjct: 362 QEKDVFEKYYKQHLAKEAFVLEKRFLMMLKERSHC*AQD*MWAINFTSKLEGMFTDMKTF 541
Query: 443 QDTMQGFYASHP 454
G P
Sbjct: 542 SGHNAGLLCQPP 577
Score = 204 bits (520), Expect = 7e-53
Identities = 103/125 (82%), Positives = 106/125 (84%), Gaps = 1/125 (0%)
Frame = +3
Query: 413 ERSLIVKLKTECGYQFTSKL-EGMFTDMKTSQDTMQGFYASHPDLGDGPTLTVQVLTTGS 471
++ LIVKLKTECG + K SQDTMQGFYASHPDLGDGPTLTVQVLTTGS
Sbjct: 450 KKGLIVKLKTECGLSILHQN*RECLQT*KPSQDTMQGFYASHPDLGDGPTLTVQVLTTGS 629
Query: 472 WPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNV 531
WPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQK ELNV
Sbjct: 630 WPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKPELNV 809
Query: 532 STYQM 536
ST M
Sbjct: 810 STLPM 824
>TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (72%)
Length = 1742
Score = 273 bits (697), Expect = 2e-73
Identities = 157/468 (33%), Positives = 262/468 (55%), Gaps = 24/468 (5%)
Frame = +3
Query: 51 LKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTPVHELGLN 110
L + S+ L EL R+W +H ++ + Y+DR FI P++E+GL
Sbjct: 345 LSTVLPSLREKHDEFMLRELVRRWANHKIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLT 524
Query: 111 LWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMRNITKMLMDLGPAV---YGQDF 167
+R+ V ++ ++ + ++ L+ ER GE IDR +++N+ + +++G Y DF
Sbjct: 525 CFRDLVY--KEVNGKVRDAVISLIDQEREGEQIDRALIKNVLDIFVEIGMGHMDHYENDF 698
Query: 168 EAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVE 227
E L+ ++ +Y ++ +I C DY+ KAE L E DRV +Y+ +E K+ + V+
Sbjct: 699 EVAMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLKREKDRVANYLHSSSEPKLLEKVQ 878
Query: 228 TQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESG 287
+++ + +L+ E+SG +L DDK EDL RM+ LF ++ GL + + H+ E G
Sbjct: 879 HELLSVYANQLLEKEHSGCHALLRDDKVEDLSRMFRLFSKIPKGLDPVSSIFKQHVTEEG 1058
Query: 288 KQLVT---------DPER-----LKDPVEFVQRLLDEKDKYDKIINQAFNNDKSFQNALN 333
LV PE+ L++ V FV+++++ DKY +N F N F AL
Sbjct: 1059TTLVKLAEDAASNKKPEKRDIVGLQEQV-FVRKVIELHDKYLAYVNDCFQNHTLFHKALK 1235
Query: 334 SSFEYFIN---LNPRSPEFISLFVDDKLRKG-LKGVNEDDVEVTLDKVMMLFRYLQEKDV 389
+FE F N S E ++ F D+ L+KG + ++++ +E TL+KV+ L Y+ +KD+
Sbjct: 1236EAFEIFCNKGVAGSSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDL 1415
Query: 390 FEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM---KTSQDTM 446
F ++Y++ LA+RLL K+ +DD ERS++ KLK +CG QFTSK+EGM TD+ K +Q +
Sbjct: 1416FAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSF 1595
Query: 447 QGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFR 494
+ + ++P++ G LTV VLTTG WP+ S NLP E+ E F+
Sbjct: 1596EEYLKNNPNVDPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 1739
>TC90868 similar to GP|9295728|gb|AAF87034.1| T24P13.25 {Arabidopsis
thaliana}, partial (22%)
Length = 779
Score = 251 bits (640), Expect = 9e-67
Identities = 124/162 (76%), Positives = 145/162 (88%)
Frame = +3
Query: 542 FNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKF 601
F+NAD+++ KEIEQAT IP DLKRCLQSLALVKG+NVLRKEPMSKD+ EDD F N+KF
Sbjct: 12 FHNADRMTCKEIEQATAIPMSDLKRCLQSLALVKGKNVLRKEPMSKDISEDDVFFFNEKF 191
Query: 602 SSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEV 661
+SKL+KVKIGTVVAQ+E+EPE ETRQRVEE+RKPQIE AIVR+MKSRR+L+HNN+IAEV
Sbjct: 192 TSKLFKVKIGTVVAQRETEPENIETRQRVEEERKPQIETAIVRVMKSRRVLEHNNVIAEV 371
Query: 662 TKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
TKQLQ RFL NP +KK+IESLIER+FLERD DR++YRYLA
Sbjct: 372 TKQLQARFLPNPVVIKKQIESLIEREFLERDKVDRELYRYLA 497
>BM813175 similar to GP|22136936|gb putative cullin 1 protein {Arabidopsis
thaliana}, partial (30%)
Length = 680
Score = 157 bits (396), Expect = 2e-38
Identities = 93/229 (40%), Positives = 134/229 (57%)
Frame = +1
Query: 464 VQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGK 523
V VLTT WP+ S NLP E+ E F+ +Y R+L+W ++G ++ F +
Sbjct: 1 VTVLTTVLWPSYKSFDLNLPSEMVKCVEVFKGFYETKTKHRKLTWIYSLGTCNIIGKF-E 177
Query: 524 GQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKE 583
+ EL VSTYQ L+LFN ADKLSY EI + DL R L SL+ K + +L KE
Sbjct: 178 PKTIELIVSTYQAAALLLFNTADKLSYSEIMTQLNLTNEDLVRLLHSLSCAKYK-ILAKE 354
Query: 584 PMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIV 643
P ++ + +D+F N KF+ K+ ++KI E+++ + V++DR+ I+AAIV
Sbjct: 355 PNTRTISPNDSFEFNSKFTDKMRRIKIPLPPVD-----ERKKVIEDVDKDRRYAIDAAIV 519
Query: 644 RIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERD 692
RIMKSR++L H L+ E +QL F + +KKRIE LI RD+LERD
Sbjct: 520 RIMKSRKVLGHQQLVLECVEQLGRMFKPDIKAIKKRIEDLITRDYLERD 666
>TC88289 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (24%)
Length = 1019
Score = 129 bits (325), Expect = 3e-30
Identities = 72/176 (40%), Positives = 111/176 (62%)
Frame = +2
Query: 528 ELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSK 587
EL V+TYQ L+LFN++D+LSY EI + D+ R L SL+ K + +L KEP +K
Sbjct: 38 ELIVTTYQASALLLFNSSDRLSYSEIMAQLNLTDDDVIRLLHSLSCAKYK-ILNKEPSTK 214
Query: 588 DVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMK 647
+ D+F N KF+ K+ ++KI EK++ + V++DR+ I+A+IVRIMK
Sbjct: 215 AILPTDSFEFNSKFTDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMK 379
Query: 648 SRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
SR++L++ L+ E +QL F + +KKRIE LI RD+LERD ++ +++YLA
Sbjct: 380 SRKVLNYQQLVMECVEQLGRMFKPDVKAIKKRIEDLISRDYLERDKDNANLFKYLA 547
>BE203271 similar to GP|22550314|gb| putative cullin protein {Olea europaea},
partial (12%)
Length = 336
Score = 48.5 bits (114), Expect = 9e-06
Identities = 23/74 (31%), Positives = 44/74 (59%), Gaps = 1/74 (1%)
Frame = +1
Query: 72 RKWNDHNKALQMIRDILMYMDRTFI-PSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTL 130
R W D + MIR I +++DRT++ S + ++GL ++R+ + S +++ + + L
Sbjct: 16 RCWQDLCDQMLMIRGIALFLDRTYVKQSPNIRSIWDMGLQIFRKHLSLSPEVQHKTVTGL 195
Query: 131 LELVQSERTGEVID 144
L ++ SER GE +D
Sbjct: 196 LRMIDSERLGEAVD 237
Score = 40.4 bits (93), Expect = 0.002
Identities = 16/32 (50%), Positives = 22/32 (68%)
Frame = +1
Query: 493 FRSYYLGTHTGRRLSWQTNMGFADLKATFGKG 524
F+ +YL ++GRRL WQ ++G LKA F KG
Sbjct: 241 FKEFYLSKYSGRRLMWQNSLGHCVLKADFSKG 336
>AJ497181 weakly similar to GP|22335691|dbj cullin-like protein1 {Pisum
sativum}, partial (10%)
Length = 461
Score = 42.7 bits (99), Expect = 5e-04
Identities = 28/130 (21%), Positives = 60/130 (45%), Gaps = 4/130 (3%)
Frame = +3
Query: 54 IAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWR 113
+ S+ + L EL +W+ H + + Y+DR FI + + E+GL +
Sbjct: 24 VLPSLRGKKDELLLRELLGRWSIHKTMTKCLSKFFHYLDRYFIGRQRLPSLEEIGLLSFY 203
Query: 114 ESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMRNITKMLMDLGPAVYGQDFEAHF-- 171
+ V ++ +++ +L ++ + GE ID ++ N ++G + G++ HF
Sbjct: 204 DLVYV--EMHREVMDAILAMIDRKWAGEPIDETLVHNALTFYSEIGEST-GKNDPKHFAE 374
Query: 172 --LQVSAEFY 179
++ +A FY
Sbjct: 375 TTIKENATFY 404
>BG450598 similar to GP|23429518|gb APC2 {Arabidopsis thaliana}, partial
(13%)
Length = 604
Score = 39.7 bits (91), Expect = 0.004
Identities = 22/101 (21%), Positives = 45/101 (43%)
Frame = +2
Query: 466 VLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQ 525
++++ WP NLP + L + + T R+L W+ ++G L+ F + +
Sbjct: 74 IISSNFWPPIQDEPLNLPEPVDKLLSDYAKRFSEVKTPRKLQWKKSLGTVKLELQF-EDR 250
Query: 526 KHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKR 566
+ + V+ ++M F + + K++ A IP L R
Sbjct: 251 EMQFTVAPVLASIIMKFQDQMSWTSKDLAAAIGIPVDVLNR 373
>BG453995 weakly similar to GP|14091836|gb| Putative cullin {Oryza sativa},
partial (6%)
Length = 670
Score = 39.7 bits (91), Expect = 0.004
Identities = 22/93 (23%), Positives = 48/93 (50%)
Frame = +2
Query: 67 LEELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRL 126
L EL +W++H +++ I Y+DR IP P E L ++Y ++++ ++
Sbjct: 383 LRELLIRWSNHKIMTKLLSRIFCYLDRFHIPRKWGLPSLEETGFLSFYHLVY-DEVKKQV 559
Query: 127 LNTLLELVQSERTGEVIDRGIMRNITKMLMDLG 159
++ +L ++ + GE ID+ ++ N ++G
Sbjct: 560 MDAILAMIDWRQAGEPIDQTLVNNALAFYSEIG 658
>TC81773 homologue to PIR|E96718|E96718 probable cullin T6C23.13 [imported]
- Arabidopsis thaliana, partial (9%)
Length = 527
Score = 38.1 bits (87), Expect = 0.012
Identities = 21/34 (61%), Positives = 24/34 (69%), Gaps = 1/34 (2%)
Frame = +1
Query: 34 KFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSF 66
KFGDRLYS + H+ K+ AKSIEAAQGG F
Sbjct: 394 KFGDRLYSRIGCDHDCHISKK*AKSIEAAQGGFF 495
Score = 32.7 bits (73), Expect = 0.49
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Frame = +3
Query: 24 FRNAYNMVLHKFG-DRLYSGLVATMTAHLKEIAKSI-EAAQGGSFLEELNRKWND 76
+RNAYNMVLH+ + + HLKEI +++ F + LNRKWND
Sbjct: 363 YRNAYNMVLHQIW**VVLKDWLRP*LPHLKEIG*IY*SSSRRVLFWKNLNRKWND 527
>TC93639 similar to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (21%)
Length = 564
Score = 36.2 bits (82), Expect = 0.044
Identities = 22/98 (22%), Positives = 43/98 (43%), Gaps = 5/98 (5%)
Frame = +1
Query: 24 FRNAYNMVLHK----FGDRLYSGLVATMTAHLKEIAKSIEAAQGGSF-LEELNRKWNDHN 78
+ YNM K + +LY +++ S + F L EL ++W +H
Sbjct: 181 YTTIYNMCTQKPPLDYSQQLYDKYKEVFDEYIRSTVLSAVRDKHDEFMLRELVQRWLNHK 360
Query: 79 KALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRESV 116
++ + Y+DR F+ P++ +GL+ +R+ V
Sbjct: 361 VLVRWLSRFFHYLDRYFVARRSLPPLNAVGLSAFRDLV 474
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 34.3 bits (77), Expect = 0.17
Identities = 34/129 (26%), Positives = 59/129 (45%), Gaps = 4/129 (3%)
Frame = +3
Query: 516 DLKATFGKGQKHELNVSTYQMCVLMLF---NNADKLSYKEIEQ-ATEIPAPDLKRCLQSL 571
++KA K QK E V+ Q + ++ +++++ S E EQ + PAP K +
Sbjct: 204 EVKAVSAKKQKVE-EVAAKQKALKVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKG 380
Query: 572 ALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVE 631
+ K + E D D V K SK K G+ A+K E++++ + +
Sbjct: 381 NVKKAQPETTSEESDSDSSSSDEEEVK-KPVSKAVPSKNGSAPAKKVDTSEEEDSEESSD 557
Query: 632 EDRKPQIEA 640
ED+KP +A
Sbjct: 558 EDKKPAAKA 584
Score = 30.0 bits (66), Expect = 3.2
Identities = 24/100 (24%), Positives = 48/100 (48%), Gaps = 3/100 (3%)
Frame = +3
Query: 545 ADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGED--DAFSVNDKFS 602
A K+ E E + E D K +++ G +K ++ ++ D ++K +
Sbjct: 507 AKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPA 686
Query: 603 SKLYKVKIGTVVAQK-ESEPEKQETRQRVEEDRKPQIEAA 641
+K K G+V A+K ++E +++ EED+KP +A+
Sbjct: 687 AKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKAS 806
>TC79996 similar to PIR|T00571|T00571 dolichyl-phosphate
beta-glucosyltransferase homolog At2g39630 - Arabidopsis
thaliana, partial (63%)
Length = 1003
Score = 32.7 bits (73), Expect = 0.49
Identities = 28/104 (26%), Positives = 45/104 (42%), Gaps = 7/104 (6%)
Frame = +3
Query: 284 RESGKQLVTDPERLKDPVEFVQ----RLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYF 339
R + Q + P +DP Q ++D KY +I A+N + AL + Y
Sbjct: 246 RRNNHQHIEVPAIFEDPNSLTQVPCPHVVDPASKYISLIIPAYNEEHRLPGALEETMIYL 425
Query: 340 INLNPRSPEFI--SLFVDDKLRKGLKGVNEDDV-EVTLDKVMML 380
+ + P F + VDD G K V + V + T+DKV ++
Sbjct: 426 QHRASKDPSFSYEVVIVDDGSVDGTKRVAFEFVRKYTVDKVRVI 557
>TC86691 similar to GP|15810175|gb|AAL06989.1 At2g03890/T18C20.9 {Arabidopsis
thaliana}, partial (76%)
Length = 2658
Score = 31.6 bits (70), Expect = 1.1
Identities = 17/36 (47%), Positives = 23/36 (63%)
Frame = -1
Query: 13 ISNFPPFCLCGFRNAYNMVLHKFGDRLYSGLVATMT 48
+S+FPP C C F + NMV F D + SG+VAT +
Sbjct: 2178 LSSFPPECFCFF--SANMVF--FSDTVISGIVATFS 2083
>AW685503 similar to GP|22165062|gb| unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 599
Score = 31.6 bits (70), Expect = 1.1
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = +2
Query: 347 PEFISLFVDDKLRKGLKGVNEDD 369
PEF+SL V +KLR+ LKG+N D
Sbjct: 248 PEFLSLPVGEKLREKLKGINTGD 316
>TC87945 weakly similar to GP|17473713|gb|AAL38309.1 unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 1087
Score = 31.2 bits (69), Expect = 1.4
Identities = 16/52 (30%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Frame = +2
Query: 127 LNTLLELVQSERT-GEVIDRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAE 177
L + L++ E T G++++ + + I + + P+ +G DFE HF Q S E
Sbjct: 455 LGPIQMLMKPESTVGDLVETAVRQYINEARRPILPSRFGSDFELHFSQFSLE 610
>TC84484 similar to GP|20161418|dbj|BAB90342. phospholipase-like protein
{Oryza sativa (japonica cultivar-group)}, partial (38%)
Length = 656
Score = 30.0 bits (66), Expect = 3.2
Identities = 15/52 (28%), Positives = 23/52 (43%)
Frame = +2
Query: 146 GIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLK 197
G M+ I + L G AV+G D+E H Y + + DC ++ K
Sbjct: 275 GFMKEIGEKLASAGYAVFGMDYEGHGHSAGVRCYITKFDNVVN--DCSNFYK 424
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,353,750
Number of Sequences: 36976
Number of extensions: 225905
Number of successful extensions: 1193
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 1157
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1170
length of query: 703
length of database: 9,014,727
effective HSP length: 103
effective length of query: 600
effective length of database: 5,206,199
effective search space: 3123719400
effective search space used: 3123719400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146568.7


