

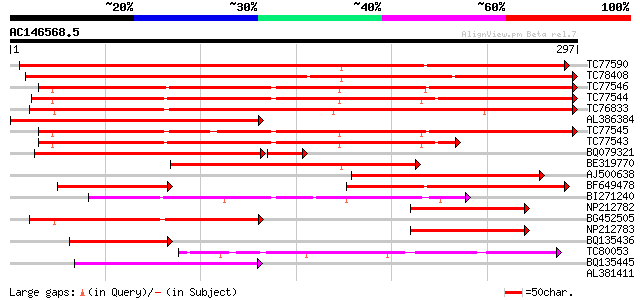
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.5 - phase: 0
(297 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77590 homologue to GP|13560120|emb|CAB65476. chitinase {Trifol... 395 e-110
TC78408 similar to GP|13560118|emb|CAB43737. chitinase {Trifoliu... 387 e-108
TC77546 similar to GP|22901738|gb|AAN10048.1 high molecular weig... 306 5e-84
TC77544 homologue to GP|22901738|gb|AAN10048.1 high molecular we... 300 6e-82
TC76833 weakly similar to SP|P51614|CHIA_VITVI Acidic endochitin... 297 4e-81
AL386384 similar to GP|10954033|gb class III acidic chitinase {M... 273 8e-74
TC77545 similar to GP|22901738|gb|AAN10048.1 high molecular weig... 266 9e-72
TC77543 homologue to GP|22901738|gb|AAN10048.1 high molecular we... 236 1e-62
BQ079321 similar to GP|10954033|gb class III acidic chitinase {M... 199 8e-56
BE319770 similar to GP|2853142|emb class III chitinase {Lupinus ... 200 5e-52
AJ500638 similar to GP|10954033|gb class III acidic chitinase {M... 174 4e-44
BF649478 homologue to GP|13560120|em chitinase {Trifolium repens... 159 2e-39
BI271240 similar to GP|22901738|gb high molecular weight root ve... 154 5e-38
NP212782 NP212782|AF167327.1|AAF67829.1 putative chitinase 132 2e-31
BG452505 weakly similar to GP|20975280|dbj chitinase {Phytolacca... 118 2e-27
NP212783 NP212783|AF167326.1|AAF67828.1 putative chitinase 111 4e-25
BQ135436 similar to GP|13560120|emb chitinase {Trifolium repens}... 65 4e-11
TC80053 similar to PIR|JC7335|JC7335 chitinase (EC 3.2.1.14) 1 -... 50 1e-06
BQ135445 44 1e-04
AL381411 homologue to GP|13560120|emb chitinase {Trifolium repen... 35 0.035
>TC77590 homologue to GP|13560120|emb|CAB65476. chitinase {Trifolium
repens}, complete
Length = 1170
Score = 395 bits (1014), Expect = e-110
Identities = 195/292 (66%), Positives = 231/292 (78%), Gaps = 4/292 (1%)
Frame = +2
Query: 6 KSTISFTFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGD 65
K++ +F LL L L S+AG IAIYWGQNGNEGTL+EACATG Y +V IAFL FG+
Sbjct: 35 KNSHTFLSLILLFLTLVATSHAGGIAIYWGQNGNEGTLSEACATGKYSHVNIAFLNKFGN 214
Query: 66 GQTPMINLAGHCDP-YSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVA 124
GQTP +NLAGHC+P N CT SS+IK CQ+KGIKVLLSIGGG GSYS++S +DA +V+
Sbjct: 215 GQTPEMNLAGHCNPSLPNSCTKFSSEIKDCQSKGIKVLLSIGGGIGSYSLSSIEDARNVS 394
Query: 125 TYLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGFN---KKVYITAAP 181
+LWN FLGGKSSSRPLG AVLDGIDFDIE GS+++W LA +LKG++ KKVY+ AAP
Sbjct: 395 KFLWNTFLGGKSSSRPLGDAVLDGIDFDIELGSTENWQHLAGFLKGYSRYGKKVYLGAAP 574
Query: 182 QCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFLG 241
QCP PD ++G AL TGLFD+VWVQFYNNPPCQYN + N ++W +W S +P KIFLG
Sbjct: 575 QCPIPDKFLGTALETGLFDFVWVQFYNNPPCQYNGN-ITNLVNSWNKWNSNVPRGKIFLG 751
Query: 242 LPASPTAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSI 293
LPAS AAGSGFI AD LTS +LPVIK S KYGGVMLWSR+ DVQ+GYS+SI
Sbjct: 752 LPASTAAAGSGFIPADVLTSEILPVIKKSRKYGGVMLWSRFHDVQTGYSTSI 907
>TC78408 similar to GP|13560118|emb|CAB43737. chitinase {Trifolium repens},
complete
Length = 1089
Score = 387 bits (993), Expect = e-108
Identities = 188/292 (64%), Positives = 230/292 (78%), Gaps = 3/292 (1%)
Frame = +1
Query: 9 ISFTFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQT 68
+S F L +++L S+A IA+YWGQNGNEG+LA+AC T NY++V IAFL TFG+GQT
Sbjct: 61 VSILLFPLFLISLFKSSHAAGIAVYWGQNGNEGSLADACNTNNYQFVNIAFLSTFGNGQT 240
Query: 69 PMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLW 128
P +NLAGHCDP SNGCT SS+I++CQ+KGIKVLLS+GGGAGSYS++S DA VA YLW
Sbjct: 241 PTLNLAGHCDPASNGCTKFSSEIQTCQSKGIKVLLSLGGGAGSYSLSSADDATQVANYLW 420
Query: 129 NNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGFN--KKVYITAAPQCPFP 186
NNFLGG SSSRPLG AVLDGIDFDIE G +H+ DLAR L GF+ ++VY++AAPQCPFP
Sbjct: 421 NNFLGGTSSSRPLGDAVLDGIDFDIEAG-GEHYDDLARALNGFSSQRRVYLSAAPQCPFP 597
Query: 187 DAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASP 246
DA + +A+ TGLFDYVWVQFYNNP CQY+ N +AW QWTS A ++FLG+PA+
Sbjct: 598 DAHLDSAINTGLFDYVWVQFYNNPQCQYSSGNTNNLVNAWNQWTSS-QAKQVFLGVPAND 774
Query: 247 TAAGS-GFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
AA S GFI +D L S VLP IKGS+KYGGVM+W R++D QSGYS++IK V
Sbjct: 775 AAAPSGGFIPSDVLISQVLPAIKGSAKYGGVMIWDRFNDGQSGYSNAIKGSV 930
>TC77546 similar to GP|22901738|gb|AAN10048.1 high molecular weight root
vegetative storage protein precursor {Medicago sativa},
partial (84%)
Length = 913
Score = 306 bits (785), Expect = 5e-84
Identities = 161/293 (54%), Positives = 199/293 (66%), Gaps = 11/293 (3%)
Frame = +3
Query: 16 LLVLAL----ANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMI 71
LLVL + S++G IAIYWGQN +GTL C TGNYE V++AFL FG G+ P
Sbjct: 18 LLVLTIFPFTIKASSSGGIAIYWGQNLGDGTLTSTCDTGNYEIVLLAFLNVFGGGRVPSW 197
Query: 72 NLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNF 131
N AGHC +S CT L +IK CQ KG+KVLLS+GG GSYS++S +DA +VA YL F
Sbjct: 198 NFAGHCGDWSP-CTKLEPEIKHCQQKGVKVLLSLGGAIGSYSLSSPEDAKNVADYLNAKF 374
Query: 132 LGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGF---NKKVYITAAPQCPFPDA 188
L G+S PLG LDGIDFDIEGG++ +W DLAR L N Y++AAPQCP PD
Sbjct: 375 LSGQSG--PLGSVTLDGIDFDIEGGTNLYWDDLARELDNLRQQNSYFYLSAAPQCPRPDH 548
Query: 189 WIGNALTTGLFDYVWVQFYNNPPCQYNP---DAFMNFEDAWKQWTSGI-PANKIFLGLPA 244
++ A+ TGLFDYV VQFYNNP CQYN DA + +W WTS + P N +F+GLPA
Sbjct: 549 YLDKAIKTGLFDYVLVQFYNNPSCQYNQANGDA-TDLLKSWNDWTSSVLPNNTVFMGLPA 725
Query: 245 SPTAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
SP AAG+G+I +DL S VLP+IK +S YGGVMLW R+ DV + YS+ IK +V
Sbjct: 726 SPDAAGNGYIPPNDLISKVLPIIKQTSNYGGVMLWDRFHDVGNDYSNQIKKYV 884
>TC77544 homologue to GP|22901738|gb|AAN10048.1 high molecular weight root
vegetative storage protein precursor {Medicago sativa},
partial (85%)
Length = 1301
Score = 300 bits (767), Expect = 6e-82
Identities = 159/300 (53%), Positives = 197/300 (65%), Gaps = 14/300 (4%)
Frame = +1
Query: 12 TFFSLLVLAL------ANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGD 65
T LLVL + S++G IA+YWGQN +G L C TGNYE V++AFL FG
Sbjct: 25 TLILLLVLTIYIFPFTIKASSSGGIAVYWGQNLGDGNLTSTCDTGNYEIVLLAFLNVFGG 204
Query: 66 GQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVAT 125
G+ P N AGHC +S CT L +IK CQ KGIKVLLSIGG +GSYS++S DA +VA
Sbjct: 205 GRVPNWNFAGHCGDWS-PCTKLEPEIKHCQQKGIKVLLSIGGASGSYSLSSPDDAKNVAD 381
Query: 126 YLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGF---NKKVYITAAPQ 182
YL+ NFL G+ PLG LDGIDFDIEGGS +W DLAR L N+ Y+ +APQ
Sbjct: 382 YLYTNFLSGQFG--PLGSVTLDGIDFDIEGGSDLYWDDLARNLDNLRQQNRYFYLASAPQ 555
Query: 183 CPFPDAWIGNALTTGLFDYVWVQFYNNPPCQY---NPDAFMNFEDAWKQWTS-GIPANKI 238
C PD ++ A+ TGLFDY+ VQFYNNPPCQY N DA + + +W WTS +P N +
Sbjct: 556 CFMPDHYLDKAIKTGLFDYILVQFYNNPPCQYDIINSDATLLLQ-SWNDWTSLALPNNTV 732
Query: 239 FLGLPASPTAAG-SGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
F+GLPA+P AA G+I DDL S VLP IK +S YGG+MLW R+ DV + YS+ IK +V
Sbjct: 733 FMGLPAAPDAAPYGGYIPPDDLISKVLPFIKPTSNYGGIMLWDRFHDVGNDYSNQIKEYV 912
>TC76833 weakly similar to SP|P51614|CHIA_VITVI Acidic endochitinase
precursor (EC 3.2.1.14). [Grape] {Vitis vinifera},
partial (70%)
Length = 1144
Score = 297 bits (760), Expect = 4e-81
Identities = 151/300 (50%), Positives = 202/300 (67%), Gaps = 13/300 (4%)
Frame = +3
Query: 11 FTFFSLLVLALA-----NGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGD 65
+++ SLL+L+++ +NAG + IYWGQ+ +G+L + C TG + V IAFL TFG
Sbjct: 27 YSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGS 206
Query: 66 GQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGG-GAGSYSIASTKDANSVA 124
G+ P +NLAGHC+P + C L I CQ++GIKV+LSIGG +YS +S +DAN +A
Sbjct: 207 GRQPQLNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLA 380
Query: 125 TYLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYL----KGFNKKVYITAA 180
Y+WNNFLGG S SRP G A+LDG+DFDIEGGS H+ LA L K KK Y+TAA
Sbjct: 381 DYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAA 560
Query: 181 PQCPFPDAWIGNALTTGLFDYVWVQFYNNP-PCQYNPDAFMNFEDAWKQWTSGIPANKIF 239
P C F D + A+ TGLFDYVWVQFYN+P C + + +F+++W QW + + K+F
Sbjct: 561 PLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVF 740
Query: 240 LGLPASPT--AAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
+GLPAS + A GF+ A DL + +LP++K S KYGGVMLW+R DV SGYSS IK+ V
Sbjct: 741 VGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 920
>AL386384 similar to GP|10954033|gb class III acidic chitinase {Malus x
domestica}, partial (39%)
Length = 428
Score = 273 bits (697), Expect = 8e-74
Identities = 133/133 (100%), Positives = 133/133 (100%)
Frame = +2
Query: 1 MKMELKSTISFTFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFL 60
MKMELKSTISFTFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFL
Sbjct: 29 MKMELKSTISFTFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFL 208
Query: 61 PTFGDGQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDA 120
PTFGDGQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDA
Sbjct: 209 PTFGDGQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDA 388
Query: 121 NSVATYLWNNFLG 133
NSVATYLWNNFLG
Sbjct: 389 NSVATYLWNNFLG 427
>TC77545 similar to GP|22901738|gb|AAN10048.1 high molecular weight root
vegetative storage protein precursor {Medicago sativa},
partial (80%)
Length = 1284
Score = 266 bits (679), Expect = 9e-72
Identities = 144/294 (48%), Positives = 189/294 (63%), Gaps = 12/294 (4%)
Frame = +2
Query: 16 LLVLAL----ANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMI 71
LLVL + S++G IAIYWGQN EGTL C T NY+ V++ FL FG G+ P +
Sbjct: 50 LLVLTIFPFTIKASSSGGIAIYWGQNVTEGTLTSTCDTDNYDIVLLTFLEFFGGGRVPSL 229
Query: 72 NLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNF 131
N AGHCD + C L +IK CQ KG KVLLS+ A ++ S+++A +++ YL+ NF
Sbjct: 230 NFAGHCDGLN--CRKLEPEIKHCQEKGFKVLLSL---AALGALNSSEEAKNLSDYLYTNF 394
Query: 132 LGGKSSSRPLGPAVLDGIDFDIEGGSSQ-HWGDLARYLKGF---NKKVYITAAPQCPFPD 187
L G+ PLG LDGIDFDIEG ++ +W DLAR L N Y++AAPQCP PD
Sbjct: 395 LSGQFG--PLGSVTLDGIDFDIEGAATNLYWDDLARELDNLRQQNSYFYLSAAPQCPMPD 568
Query: 188 AWIGNALTTGLFDYVWVQFYNNPPCQY---NPDAFMNFEDAWKQWTSGI-PANKIFLGLP 243
++ A+ TGLFDY+ VQFY+N PCQY N DA + +W WTS + P N +F+GLP
Sbjct: 569 YYLDKAIKTGLFDYILVQFYHNTPCQYDQINSDA-TDLLKSWNAWTSSVLPNNTVFMGLP 745
Query: 244 ASPTAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
ASP AA G+I DDL S VLP+IK +S YGGV+L R D+++ YS+ IK +V
Sbjct: 746 ASPDAASEGYIPPDDLISKVLPIIKQTSNYGGVVL*DRAHDIENDYSNQIKEYV 907
>TC77543 homologue to GP|22901738|gb|AAN10048.1 high molecular weight root
vegetative storage protein precursor {Medicago sativa},
partial (70%)
Length = 737
Score = 236 bits (601), Expect = 1e-62
Identities = 126/232 (54%), Positives = 154/232 (66%), Gaps = 11/232 (4%)
Frame = +3
Query: 16 LLVLAL----ANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMI 71
LLVL + S++G IAIYWGQN +GTL C TGNYE V++AFL FG G+ P
Sbjct: 51 LLVLTIFPFTIKASSSGGIAIYWGQNLGDGTLTSTCDTGNYEIVLLAFLNVFGGGRVPSW 230
Query: 72 NLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNF 131
N AGHC +S CT L +IK CQ KGIKVLLS+GG +GSYS++S DA +VA YL+ NF
Sbjct: 231 NFAGHCGDWS-PCTKLEPEIKHCQQKGIKVLLSLGGASGSYSLSSPDDAKNVADYLYTNF 407
Query: 132 LGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGF---NKKVYITAAPQCPFPDA 188
L G+ PLG LDGIDFDIEGGS+ +W DLAR L N+ Y+ +APQC PD
Sbjct: 408 LSGQFG--PLGSVTLDGIDFDIEGGSNLYWDDLARNLDNLRQQNRYFYLASAPQCFMPDY 581
Query: 189 WIGNALTTGLFDYVWVQFYNNPPCQY---NPDAFMNFEDAWKQWTS-GIPAN 236
++ A+ TGLFDYV VQFYNNPPCQY N DA + + +W WTS +P N
Sbjct: 582 YLDKAIKTGLFDYVLVQFYNNPPCQYDLINSDATLLLQ-SWNAWTSLALPNN 734
>BQ079321 similar to GP|10954033|gb class III acidic chitinase {Malus x
domestica}, partial (47%)
Length = 552
Score = 199 bits (505), Expect(2) = 8e-56
Identities = 93/121 (76%), Positives = 106/121 (86%)
Frame = +2
Query: 14 FSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMINL 73
FSL++L LA GSN G I+IYWGQNGNEGTLAE CATGNYEYV IAFL TFG+G+ P ++L
Sbjct: 65 FSLVILILARGSNGGGISIYWGQNGNEGTLAETCATGNYEYVNIAFLYTFGNGRVPRMDL 244
Query: 74 AGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNFLG 133
AGHCD YSNGCT +S+DIKSCQ+KGIKV+LSIGGGAGSYS+AS +DA ATYLWNNFLG
Sbjct: 245 AGHCDAYSNGCTKISADIKSCQSKGIKVILSIGGGAGSYSLASLEDARIGATYLWNNFLG 424
Query: 134 G 134
G
Sbjct: 425 G 427
Score = 35.8 bits (81), Expect(2) = 8e-56
Identities = 17/21 (80%), Positives = 18/21 (84%)
Frame = +3
Query: 136 SSSRPLGPAVLDGIDFDIEGG 156
SS+R LG A LDGIDFDIEGG
Sbjct: 432 SSTRXLGDAGLDGIDFDIEGG 494
>BE319770 similar to GP|2853142|emb class III chitinase {Lupinus albus},
partial (46%)
Length = 437
Score = 200 bits (509), Expect = 5e-52
Identities = 93/133 (69%), Positives = 111/133 (82%), Gaps = 2/133 (1%)
Frame = +2
Query: 85 TGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNFLGGKSSSRPLGPA 144
T S DI++CQ+KGIKVLLSIGGG GSYS+ S+ DA+ +A YLWNNFLGG SSSRPLG A
Sbjct: 2 TNFSKDIQTCQSKGIKVLLSIGGGTGSYSLYSSYDASQLANYLWNNFLGGTSSSRPLGDA 181
Query: 145 VLDGIDFDIEGGSSQHWGDLARYLKGFN--KKVYITAAPQCPFPDAWIGNALTTGLFDYV 202
VLDGIDFDIE G Q+W +LA+ L GF+ KKVY++AAPQCP+PDA + +A+ TGLFDYV
Sbjct: 182 VLDGIDFDIEAGDGQYWDELAKALDGFSSQKKVYLSAAPQCPYPDAHLDSAIKTGLFDYV 361
Query: 203 WVQFYNNPPCQYN 215
WVQFYNNP CQY+
Sbjct: 362 WVQFYNNPQCQYS 400
>AJ500638 similar to GP|10954033|gb class III acidic chitinase {Malus x
domestica}, partial (33%)
Length = 307
Score = 174 bits (441), Expect = 4e-44
Identities = 83/101 (82%), Positives = 84/101 (82%)
Frame = -3
Query: 180 APQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIF 239
APQCPFPDAWIG AL TGLFD VWVQFYNNPPCQY+ A N EDAWKQ S IPA KIF
Sbjct: 305 APQCPFPDAWIGKALKTGLFDCVWVQFYNNPPCQYSGGAISNLEDAWKQRISDIPAKKIF 126
Query: 240 LGLPASPTAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWS 280
LGLPASP AAGSGFISA DLTS VLP IK SSKYGGVMLWS
Sbjct: 125 LGLPASPQAAGSGFISATDLTSKVLPAIKDSSKYGGVMLWS 3
>BF649478 homologue to GP|13560120|em chitinase {Trifolium repens}, partial
(58%)
Length = 633
Score = 159 bits (401), Expect = 2e-39
Identities = 76/117 (64%), Positives = 89/117 (75%)
Frame = +1
Query: 177 ITAAPQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPAN 236
IT+ PD ++G AL TGLFD+VWVQFYNNPPCQYN + N ++W +W S +P
Sbjct: 163 ITSKFMHKIPDKFLGTALETGLFDFVWVQFYNNPPCQYNGN-ITNLVNSWNKWNSNVPRG 339
Query: 237 KIFLGLPASPTAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSI 293
KIFLGLPAS AAGSGFI AD LTS +LPVIK S KYGGVMLWSR+ DVQ+GYS+SI
Sbjct: 340 KIFLGLPASTAAAGSGFIPADVLTSEILPVIKKSRKYGGVMLWSRFHDVQTGYSTSI 510
Score = 94.4 bits (233), Expect = 5e-20
Identities = 44/61 (72%), Positives = 50/61 (81%), Gaps = 1/61 (1%)
Frame = +2
Query: 26 NAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMINLAGHCDP-YSNGC 84
+AG IAIYWGQNGNEGTL+EACATG Y +V IAFL FG+GQTP +NLAGHC+P N C
Sbjct: 2 HAGGIAIYWGQNGNEGTLSEACATGKYSHVNIAFLNKFGNGQTPEMNLAGHCNPSLPNSC 181
Query: 85 T 85
T
Sbjct: 182T 184
>BI271240 similar to GP|22901738|gb high molecular weight root vegetative
storage protein precursor {Medicago sativa}, partial
(40%)
Length = 669
Score = 154 bits (388), Expect = 5e-38
Identities = 90/212 (42%), Positives = 121/212 (56%), Gaps = 12/212 (5%)
Frame = +3
Query: 42 TLAEACATGNYEYVIIAFLPTFGDGQT-PMINLAGHCDPYSNGCTGLSSDIKSCQAKGIK 100
TL C TG Y+ V+++FL F +G+ P +N GHC+ N CT L +I CQ KG+K
Sbjct: 3 TLKSTCDTGFYKIVLLSFLNIFQEGRRIPKLNFIGHCND-KNPCTNLEPEIIHCQQKGVK 179
Query: 101 VLLSIGGGAGS--YSIASTKDANSVATYLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSS 158
V LS+GG + YS+ S +DA +VA YL+ NFL G+ PLG L+GI DI+GGS
Sbjct: 180 VFLSLGGAYENETYSLGSLEDAKNVANYLFTNFLNGQFG--PLGSVTLNGISLDIQGGSD 353
Query: 159 QHWGDLARYLKGFNKKV---YITAAPQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYN 215
Q W A+YL + YI A QC PD ++ A+ +G FDY V+FYNNP CQY+
Sbjct: 354 Q-WEFFAKYLLYVRQNYRYFYIAVASQCIIPDQYLDKAIKSGSFDYAIVKFYNNPNCQYD 530
Query: 216 PDAFMNFED-----AWKQWTSGIPA-NKIFLG 241
NF D +W WT + N +F+G
Sbjct: 531 Q---TNFNDTLLTRSWNSWTWLVQLDNNVFMG 617
>NP212782 NP212782|AF167327.1|AAF67829.1 putative chitinase
Length = 188
Score = 132 bits (332), Expect = 2e-31
Identities = 62/62 (100%), Positives = 62/62 (100%)
Frame = +2
Query: 211 PCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASPTAAGSGFISADDLTSTVLPVIKGS 270
PCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASPTAAGSGFISADDLTSTVLPVIKGS
Sbjct: 2 PCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASPTAAGSGFISADDLTSTVLPVIKGS 181
Query: 271 SK 272
SK
Sbjct: 182 SK 187
>BG452505 weakly similar to GP|20975280|dbj chitinase {Phytolacca americana},
partial (25%)
Length = 669
Score = 118 bits (296), Expect = 2e-27
Identities = 59/129 (45%), Positives = 82/129 (62%), Gaps = 6/129 (4%)
Frame = +2
Query: 11 FTFFSLLVLALA-----NGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGD 65
+++ SLL+L+++ +NAG + IYWGQ +G+L + C TG + V IAFL TFG
Sbjct: 278 YSYPSLLLLSISLTLFFQSTNAGXLVIYWGQXNGDGSLXDTCNTGLFNIVNIAFLSTFGS 457
Query: 66 GQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGG-GAGSYSIASTKDANSVA 124
G+ P +NLAGHC+P C L I C + IKV+LSIGG +YS +S +DAN +A
Sbjct: 458 GRQPQLNLAGHCNP--PNCKNLRDSINICXSXXIKVMLSIGGEXRNTYSFSSPEDANQLA 631
Query: 125 TYLWNNFLG 133
Y+WNNF G
Sbjct: 632 DYIWNNFFG 658
>NP212783 NP212783|AF167326.1|AAF67828.1 putative chitinase
Length = 188
Score = 111 bits (277), Expect = 4e-25
Identities = 52/62 (83%), Positives = 53/62 (84%)
Frame = +2
Query: 211 PCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASPTAAGSGFISADDLTSTVLPVIKGS 270
PCQYNP N EDAWKQWTSGIPANKIFLGLPASP AAGSGFI A DLTSTVLP IKGS
Sbjct: 2 PCQYNPGEISNLEDAWKQWTSGIPANKIFLGLPASPEAAGSGFIPATDLTSTVLPAIKGS 181
Query: 271 SK 272
+K
Sbjct: 182 AK 187
>BQ135436 similar to GP|13560120|emb chitinase {Trifolium repens}, partial
(21%)
Length = 746
Score = 64.7 bits (156), Expect = 4e-11
Identities = 33/55 (60%), Positives = 38/55 (69%), Gaps = 1/55 (1%)
Frame = +3
Query: 32 IYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMINLAGHCDP-YSNGCT 85
IYWGQNGN+GTL+EAC T Y +V I+ L DGQT +NLA HC P SN CT
Sbjct: 123 IYWGQNGNQGTLSEACPT*KYFHVNISTLNKNSDGQTHELNLACHCYPSLSNTCT 287
>TC80053 similar to PIR|JC7335|JC7335 chitinase (EC 3.2.1.14) 1 - cone shell
(Conus tulipa), partial (81%)
Length = 1081
Score = 49.7 bits (117), Expect = 1e-06
Identities = 59/213 (27%), Positives = 90/213 (41%), Gaps = 12/213 (5%)
Frame = +2
Query: 89 SDIKSCQAKGIKVLLSIGGGA--GSYSIASTKDANSVATYLWNNFLGGKSSSRPLGPAVL 146
S IKS Q +KV LS+GG + G Y+ D +SV ++L N S ++ + L
Sbjct: 350 SSIKS-QNPNVKVALSLGGDSVEGGYAYF---DPSSVESWLSNAV---SSLTKIIKEYNL 508
Query: 147 DGIDFDIE---GGSSQHWGDLARYLKGFNKKVYITAAPQCPFPDAWIGNALTT------G 197
DGID D E G + + R +K IT A PF D + +
Sbjct: 509 DGIDIDYEHFKGNPNTFAECIGRLIKTLKANGVITFASIAPFDDDQVQSHYLALWKSYGH 688
Query: 198 LFDYVWVQFYNNPPCQYNPDAFMN-FEDAWKQWTSGIPANKIFLGLPASPTAAGSGFISA 256
L DYV QFY Y+ ++ F D + + +S K+ + + GSG +S
Sbjct: 689 LIDYVNFQFY-----AYDKGTSVSQFIDYFNKQSSNYNGGKVLVSFLSD----GSGGLSP 841
Query: 257 DDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGY 289
D +K K G+ +WS D + +G+
Sbjct: 842 SDGFFKACQRLKSQQKLHGIFVWSADDSMGNGF 940
>BQ135445
Length = 758
Score = 43.5 bits (101), Expect = 1e-04
Identities = 28/99 (28%), Positives = 42/99 (42%), Gaps = 1/99 (1%)
Frame = +2
Query: 35 GQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMINLAGHCDPY-SNGCTGLSSDIKS 93
G GNEG L+ C T Y V + L + G GQ +NL + P+ ++ T ++ I
Sbjct: 122 GLEGNEGPLSHPCPTEKYNPVHMTSLKSNGSGQITEMNLTSNGIPFLTHSFTPITFHISD 301
Query: 94 CQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNFL 132
C +LS+ Y + T DA + N L
Sbjct: 302 CHTYATTFVLSVTASIEHYFLLLTGDAQTSLNIFQRNHL 418
>AL381411 homologue to GP|13560120|emb chitinase {Trifolium repens}, partial
(8%)
Length = 151
Score = 35.0 bits (79), Expect = 0.035
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = +2
Query: 106 GGGAGSYSIASTKDANSVATYLWNNFLG 133
G GSYS++S +DA +V+ +LWN F G
Sbjct: 59 GCRVGSYSLSSIEDARNVSKFLWNTFYG 142
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,391,788
Number of Sequences: 36976
Number of extensions: 156786
Number of successful extensions: 693
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 652
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 657
length of query: 297
length of database: 9,014,727
effective HSP length: 95
effective length of query: 202
effective length of database: 5,502,007
effective search space: 1111405414
effective search space used: 1111405414
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146568.5


