

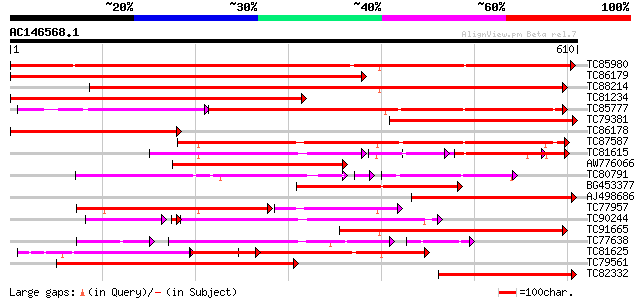
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.1 + phase: 0
(610 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85980 similar to GP|15294146|gb|AAK95250.1 AT4g18030/T6K21_210... 855 0.0
TC86179 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 812 0.0
TC88214 similar to GP|4325339|gb|AAD17339.1| F15P23.1 gene produ... 694 0.0
TC81234 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 606 e-174
TC85777 similar to PIR|F86442|F86442 unknown protein [imported] ... 358 e-137
TC79381 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 414 e-116
TC86178 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 398 e-111
TC87587 similar to GP|20259233|gb|AAM14332.1 putative ankyrin pr... 361 e-100
TC81615 similar to GP|11994314|dbj|BAB02273. ankyrin-like protei... 214 5e-97
AW776066 similar to GP|6752888|gb| unknown {Malus x domestica}, ... 327 6e-90
TC80791 similar to GP|16648931|gb|AAL24317.1 Unknown protein {Ar... 202 2e-83
BG453377 similar to GP|6752888|gb| unknown {Malus x domestica}, ... 303 1e-82
AJ498686 similar to GP|6752888|gb| unknown {Malus x domestica}, ... 296 2e-80
TC77957 similar to GP|15450900|gb|AAK96721.1 Unknown protein {Ar... 223 2e-76
TC90244 similar to GP|15450749|gb|AAK96646.1 At2g39750/T5I7.5 {A... 213 6e-76
TC91665 similar to PIR|T02472|T02472 hypothetical protein At2g45... 280 1e-75
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 188 2e-69
TC81625 similar to PIR|T02472|T02472 hypothetical protein At2g45... 250 1e-66
TC79561 similar to GP|14194107|gb|AAK56248.1 At1g77260/T14N5_19 ... 239 2e-63
TC82332 similar to GP|16604525|gb|AAL24268.1 AT4g00750/F15P23_1 ... 209 2e-54
>TC85980 similar to GP|15294146|gb|AAK95250.1 AT4g18030/T6K21_210
{Arabidopsis thaliana}, partial (82%)
Length = 2287
Score = 855 bits (2209), Expect = 0.0
Identities = 403/618 (65%), Positives = 491/618 (79%), Gaps = 11/618 (1%)
Frame = +3
Query: 2 AKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNA--ECDVVP 59
+K + NR R + IF V+ LCC FY+LG WQ SG GKGDS+AL++ K A +C++VP
Sbjct: 327 SKNALPGNRARRPLSIFAVLALCCLFYLLGTWQSSGSGKGDSLALKVNKMQATTDCNIVP 506
Query: 60 NLSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCP 119
NL+F+ H V +++S K+FK C+ +Y DYTPCQ+Q RAMTFPRENM YRERHCP
Sbjct: 507 NLNFEPQHK-YVEIVEQSEPKAKMFKACDVKYADYTPCQEQDRAMTFPRENMIYRERHCP 683
Query: 120 PEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPG 179
P+EEKL C+IPAP+GY TPFPWPKSRDY YAN PYK LTVEKA QNW+++EGNVF+FPG
Sbjct: 684 PQEEKLRCLIPAPEGYTTPFPWPKSRDYAYYANVPYKHLTVEKANQNWVKFEGNVFKFPG 863
Query: 180 GGTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSH 239
GGT FPQGAD YID+LASVIPI DGTVRTALDTGCGVASWGAYL RNV+AMSFAP+D+H
Sbjct: 864 GGTMFPQGADAYIDELASVIPIKDGTVRTALDTGCGVASWGAYLLKRNVLAMSFAPKDNH 1043
Query: 240 EAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRP 299
EAQVQFALERGVPA+IGV G+I+LP+PSRAFDM+ CSRCLIPW N+GMY+MEVDRVLRP
Sbjct: 1044EAQVQFALERGVPAIIGVLGSIRLPFPSRAFDMSQCSRCLIPWAENEGMYLMEVDRVLRP 1223
Query: 300 GGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTD 359
GG+W+LSGPPINWK Y+ W+R KE+L+ EQ++IE++A+ LCWEKK EK +IAIW+K +
Sbjct: 1224GGFWILSGPPINWKTYYQTWKRTKEDLKAEQKRIEDLAESLCWEKKYEKGDIAIWRKKIN 1403
Query: 360 TESCRSRQDDSSVEFCESSDPDDVWYKKLKACVTPTP------KVSGGDLKPFPDRLYAI 413
+SC+ + S+ C+ + DDVWYKK++ C TP P +V+GG+LK FP RL+A+
Sbjct: 1404AKSCQRK----SLNLCDLDNADDVWYKKMEVCKTPIPEVTSQSEVAGGELKKFPARLFAV 1571
Query: 414 PPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAI 473
PPR++ +IPGV+ E+YQ DNK+WKK VN YK+IN L+ + RYRNIMDMNA LG FAAA+
Sbjct: 1572PPRIAKDAIPGVTVESYQEDNKLWKKRVNNYKRINRLIGTTRYRNIMDMNARLGGFAAAL 1751
Query: 474 HSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQD 533
S KSWVMN VPTIA+ +TLGVIYERGLIGIYHDWCEGF+TYP TYDLIHANGLFSLY D
Sbjct: 1752ESQKSWVMNAVPTIAD-NTLGVIYERGLIGIYHDWCEGFATYPXTYDLIHANGLFSLYXD 1928
Query: 534 KCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVK-KLIGGMRWNMKLVDHEDGPL-VPEK 591
KCN +DILLEMDRILRPEG+VIIRDE+DV+ KV+ G RW K+VD P+ +
Sbjct: 1929KCNLQDILLEMDRILRPEGSVIIRDEIDVINKVQ*NCSGEWRWESKMVDS*RXPICTXXR 2108
Query: 592 VLIAVKQYWV-TDGNSTS 608
+ K YWV N+TS
Sbjct: 2109YWLL*KNYWVGCSKNNTS 2162
>TC86179 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (60%)
Length = 1297
Score = 812 bits (2097), Expect = 0.0
Identities = 383/383 (100%), Positives = 383/383 (100%)
Frame = +1
Query: 1 MAKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPN 60
MAKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPN
Sbjct: 148 MAKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPN 327
Query: 61 LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP 120
LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP
Sbjct: 328 LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP 507
Query: 121 EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG 180
EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG
Sbjct: 508 EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG 687
Query: 181 GTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHE 240
GTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHE
Sbjct: 688 GTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHE 867
Query: 241 AQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPG 300
AQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPG
Sbjct: 868 AQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPG 1047
Query: 301 GYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDT 360
GYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDT
Sbjct: 1048GYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDT 1227
Query: 361 ESCRSRQDDSSVEFCESSDPDDV 383
ESCRSRQDDSSVEFCESSDPDDV
Sbjct: 1228ESCRSRQDDSSVEFCESSDPDDV 1296
Score = 36.6 bits (83), Expect = 0.029
Identities = 26/103 (25%), Positives = 47/103 (45%), Gaps = 1/103 (0%)
Frame = +1
Query: 451 LDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCE 510
++ G R +D G+ S+ A + S M+ P + ++ + ERG+ + +
Sbjct: 748 INDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGT 927
Query: 511 GFSTYP-RTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEG 552
YP R +D+ H + L N ++E+DR+LRP G
Sbjct: 928 IKLPYPSRAFDMAHCSRC--LIPWGANDGMYMMEVDRVLRPGG 1050
>TC88214 similar to GP|4325339|gb|AAD17339.1| F15P23.1 gene product
{Arabidopsis thaliana}, partial (83%)
Length = 1866
Score = 694 bits (1791), Expect = 0.0
Identities = 315/522 (60%), Positives = 406/522 (77%), Gaps = 8/522 (1%)
Frame = +1
Query: 87 CEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLHCMIPAPKGYVTPFPWPKSRD 146
C+ ++YTPC+D +R++ FPREN+ YRERHCP +EE L C IPAP GY P WP+SRD
Sbjct: 34 CDVALSEYTPCEDTQRSLKFPRENLIYRERHCPEKEEVLRCRIPAPYGYRVPPRWPESRD 213
Query: 147 YVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIPINDGTV 206
+ YAN P+K LT+EK QNW+ +EG+ FRFPGGGT FP+GA YID + +I + DG+V
Sbjct: 214 WAWYANVPHKELTIEKKNQNWVHFEGDRFRFPGGGTMFPRGAGAYIDDIGKLINLKDGSV 393
Query: 207 RTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYP 266
RTALDTGCGVASWGAYL R+++A+SFAPRD+HEAQVQFALERGVPA+IGV +I+LPYP
Sbjct: 394 RTALDTGCGVASWGAYLLPRDILAVSFAPRDTHEAQVQFALERGVPALIGVIASIRLPYP 573
Query: 267 SRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEEL 326
SRAFDMAHCSRCLIPWG NDG+Y+ EVDRVLRPGGYW+LSGPPINW+ ++K W+R +E+L
Sbjct: 574 SRAFDMAHCSRCLIPWGQNDGIYLTEVDRVLRPGGYWILSGPPINWESHWKGWERTREDL 753
Query: 327 EEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCR-SRQDDSSVEFCESSDPDDVWY 385
EQ IE VAK LCW+K +K +IAIWQK T+ C+ +R+ + FC++ DPD WY
Sbjct: 754 NAEQTSIERVAKSLCWKKLVQKGDIAIWQKPTNHIHCKITRKVFKNRPFCDAKDPDSAWY 933
Query: 386 KKLKACVTPTP------KVSGGDLKPFPDRLYAIPPRVSSGSIPGVSSETYQNDNKMWKK 439
K+ C+TP P +VSG L +P+RL ++PPR+SSGS+ G+++E ++ + ++WKK
Sbjct: 934 TKMDTCLTPLPEVTDIKEVSGRGLSNWPERLTSVPPRISSGSLDGITAEMFKENTELWKK 1113
Query: 440 HVNAYKKIN-SLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYE 498
V YK ++ L + GRYRN++DMNA LG FAAA+ WVMNVVP AE +TLGV+YE
Sbjct: 1114RVAYYKTLDYQLAEPGRYRNLLDMNAYLGGFAAAMIDDPVWVMNVVPVEAEINTLGVVYE 1293
Query: 499 RGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVIIRD 558
RGLIG Y +WCE STYPRTYD IHA+ LF+LY+D+CN EDIL+EMDRILRP+G+VI+RD
Sbjct: 1294RGLIGTYQNWCEAMSTYPRTYDFIHADSLFTLYEDRCNIEDILVEMDRILRPQGSVILRD 1473
Query: 559 EVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYW 600
+VDVL+KVK+ M+W+ ++ DHE GP EK+L+AVKQYW
Sbjct: 1474DVDVLLKVKRFADAMQWDARIADHEKGPHQREKILVAVKQYW 1599
>TC81234 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (50%)
Length = 1149
Score = 606 bits (1563), Expect = e-174
Identities = 278/319 (87%), Positives = 297/319 (92%)
Frame = +3
Query: 1 MAKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPN 60
MAKPS++ +RTRS VQIFIVVGLCCFFYILGAWQR+GFGKGD + LE+TK A CD+VPN
Sbjct: 192 MAKPSSSGSRTRSFVQIFIVVGLCCFFYILGAWQRTGFGKGDLLQLEVTKKGAGCDIVPN 371
Query: 61 LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP 120
LSFDSHH GEV +IDE +S KVFKPC+ARY DYTPC DQRRAMTF R+NM YRERHCP
Sbjct: 372 LSFDSHHGGEVGKIDEVDSKPKVFKPCKARYIDYTPCHDQRRAMTFSRQNMIYRERHCPR 551
Query: 121 EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG 180
EEEKLHC+IPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG
Sbjct: 552 EEEKLHCLIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG 731
Query: 181 GTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHE 240
GTQFPQGADKYIDQ+ASVIPI +GTVRTALDTGCGVASWGAYLWSRNV+AMSFAPRDSHE
Sbjct: 732 GTQFPQGADKYIDQIASVIPIENGTVRTALDTGCGVASWGAYLWSRNVIAMSFAPRDSHE 911
Query: 241 AQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPG 300
AQVQFALERGVPAVIGV GTIKLPYPS AFDMAHCSRCLIPWG+NDG+Y+MEVDRVLRPG
Sbjct: 912 AQVQFALERGVPAVIGVLGTIKLPYPSGAFDMAHCSRCLIPWGSNDGIYLMEVDRVLRPG 1091
Query: 301 GYWVLSGPPINWKVNYKPW 319
GYWVLSGPPI+WK NYK W
Sbjct: 1092GYWVLSGPPIHWKANYKAW 1148
Score = 34.3 bits (77), Expect = 0.14
Identities = 25/103 (24%), Positives = 46/103 (44%), Gaps = 1/103 (0%)
Frame = +3
Query: 451 LDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCE 510
+++G R +D G+ S+ A + S M+ P + ++ + ERG+ +
Sbjct: 792 IENGTVRTALDTGCGVASWGAYLWSRNVIAMSFAPRDSHEAQVQFALERGVPAVIGVLGT 971
Query: 511 GFSTYPR-TYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEG 552
YP +D+ H + + N L+E+DR+LRP G
Sbjct: 972 IKLPYPSGAFDMAHCSRCLIPWGS--NDGIYLMEVDRVLRPGG 1094
>TC85777 similar to PIR|F86442|F86442 unknown protein [imported] - Arabidopsis
thaliana, partial (89%)
Length = 2381
Score = 358 bits (919), Expect(2) = e-137
Identities = 177/391 (45%), Positives = 246/391 (62%), Gaps = 5/391 (1%)
Frame = +2
Query: 215 GVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAH 274
GVASWG L R V+ +S APRD+HEAQVQFALERG+PA++GV T +LP+PS +FDMAH
Sbjct: 755 GVASWGGDLLDRGVLTISLAPRDNHEAQVQFALERGIPAILGVISTQRLPFPSNSFDMAH 934
Query: 275 CSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIE 334
CSRCLIPW G+Y+ E+ R+LRPGG+WVLSGPP+N++ ++ W EE + K++
Sbjct: 935 CSRCLIPWTEFGGIYLQEIHRILRPGGFWVLSGPPVNYERRWRGWNTTVEEQRTDYEKLQ 1114
Query: 335 EVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVEFCESS-DPDDVWYKKLKAC-V 392
++ +C++ ++K +I +WQK D D+ C+ S +PD WY L+AC V
Sbjct: 1115 DLLTSMCFKLYNKKDDIYVWQKAKDNACYDKLSRDTYPPKCDDSLEPDSAWYTPLRACFV 1294
Query: 393 TPTPKVSGGDL---KPFPDRLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINS 449
P K L +P RL P R+S + G SS T+ +DN WKK + YKK+
Sbjct: 1295 VPMEKYKKSGLTYMPKWPQRLNVAPERIS--LVQGSSSSTFSHDNSKWKKRIQHYKKLLP 1468
Query: 450 LLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWC 509
L + + RN+MDMN G FAA++ + WVMNVV + +TL V+++RGLIG +HDWC
Sbjct: 1469 DLGTNKIRNVMDMNTAYGGFAASLINDPLWVMNVVSSYG-PNTLPVVFDRGLIGTFHDWC 1645
Query: 510 EGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKL 569
E FSTYPRTYDL+HA+G F+ +C + ++LEMDRILRP G IIR+ + +
Sbjct: 1646 EAFSTYPRTYDLLHADGFFTAESHRCEMKYVMLEMDRILRPGGHAIIRESSYFADAIATM 1825
Query: 570 IGGMRWNMKLVDHEDGPLVPEKVLIAVKQYW 600
GMRW + E G + EK+L+ K+ W
Sbjct: 1826 AKGMRWICHKENTEFG-VEKEKILVCQKKLW 1915
Score = 147 bits (372), Expect(2) = e-137
Identities = 82/210 (39%), Positives = 108/210 (51%), Gaps = 3/210 (1%)
Frame = +1
Query: 9 NRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPNL--SFDSH 66
N+ RS ++ LC F + LG +SG N DV+ + S DS
Sbjct: 181 NKNRSLTAAITIIVLCGFSFYLGGVFKSG--------------NNGVDVINTIQKSLDSP 318
Query: 67 HAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLH 126
S + S F C Y DYTPC D +R + + ERHCPP E+
Sbjct: 319 KQSSGSLQIKPFS----FPECSNDYQDYTPCTDPKRWRKYGTYRLTLLERHCPPIFERKE 486
Query: 127 CMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQ 186
C++P P GY P WPKSRD Y N PY + +K+ Q+W+ EG F+FPGGGT FP
Sbjct: 487 CLVPPPPGYKPPIRWPKSRDECWYRNVPYDWINKQKSNQHWLIKEGEKFQFPGGGTMFPN 666
Query: 187 GADKYIDQLASVIP-INDGTVRTALDTGCG 215
G +Y+D + +IP I DG+VRTA+DTGCG
Sbjct: 667 GVGEYVDLMQDLIPGIKDGSVRTAIDTGCG 756
>TC79381 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (31%)
Length = 981
Score = 414 bits (1065), Expect = e-116
Identities = 202/202 (100%), Positives = 202/202 (100%)
Frame = +2
Query: 409 RLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGS 468
RLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGS
Sbjct: 8 RLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGS 187
Query: 469 FAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLF 528
FAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLF
Sbjct: 188 FAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLF 367
Query: 529 SLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLV 588
SLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLV
Sbjct: 368 SLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLV 547
Query: 589 PEKVLIAVKQYWVTDGNSTSTQ 610
PEKVLIAVKQYWVTDGNSTSTQ
Sbjct: 548 PEKVLIAVKQYWVTDGNSTSTQ 613
Score = 36.6 bits (83), Expect = 0.029
Identities = 26/103 (25%), Positives = 47/103 (45%), Gaps = 2/103 (1%)
Frame = +2
Query: 201 INDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGT 260
++ G R +D G+ S+ A + S M+ P + ++ + ERG+ + +
Sbjct: 134 LDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCE 313
Query: 261 IKLPYPSRAFDMAHCSRC--LIPWGANDGMYMMEVDRVLRPGG 301
YP R +D+ H + L N ++E+DR+LRP G
Sbjct: 314 GFSTYP-RTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEG 439
>TC86178 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (29%)
Length = 691
Score = 398 bits (1023), Expect = e-111
Identities = 185/185 (100%), Positives = 185/185 (100%)
Frame = +3
Query: 1 MAKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPN 60
MAKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPN
Sbjct: 135 MAKPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPN 314
Query: 61 LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP 120
LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP
Sbjct: 315 LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP 494
Query: 121 EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG 180
EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG
Sbjct: 495 EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG 674
Query: 181 GTQFP 185
GTQFP
Sbjct: 675 GTQFP 689
>TC87587 similar to GP|20259233|gb|AAM14332.1 putative ankyrin protein
{Arabidopsis thaliana}, partial (70%)
Length = 1641
Score = 361 bits (926), Expect = e-100
Identities = 191/441 (43%), Positives = 274/441 (61%), Gaps = 19/441 (4%)
Frame = +3
Query: 181 GTQFPQGADKYIDQLASVIPI------NDGTVRTALDTGCGVASWGAYLWSRNVVAMSFA 234
GT F GADKYI +A+++ N+G +RT LD GCGVAS+GAYL + +++AMS A
Sbjct: 24 GTHFHHGADKYIAAIANMLNFTDNNLNNEGRLRTVLDVGCGVASFGAYLLASDIIAMSLA 203
Query: 235 PRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVD 294
P D H+ Q+QFALERG+PA +GV GT +LPYPSR+F++AHCSRC I W DG+ ++E+D
Sbjct: 204 PNDVHQNQIQFALERGIPAYLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELD 383
Query: 295 RVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIW 354
RVLRPGGY+ S P + + + +E+L R++ + ++CW S++ + IW
Sbjct: 384 RVLRPGGYFAYSSP--------EAYAQDEEDL-RIWREMSALVGRMCWRIASKRDQTVIW 536
Query: 355 QKMTDTESCRSRQDDSSVEFCES-SDPDDVWYKKLKACVTP----TPKVSGGDLKPFPDR 409
QK + R+ + C+S DPD VW ++ C+TP + G +L P+P R
Sbjct: 537 QKPLTNDCYMEREPGTRPPLCQSDEDPDAVWGVNMEVCITPYSDHDNRAQGSELAPWPSR 716
Query: 410 LYAIPPRVSSGSIPGVSSETYQNDNKMWKKHV-NAYKKINSLLDSGRYRNIMDMNAGLGS 468
L + PPR++ G+SSE ++ D ++W+K V N + + + S RN+MDM A LGS
Sbjct: 717 LTSPPPRLAD---LGLSSEVFEKDTELWQKRVENYWNLMGPKISSNTVRNVMDMKANLGS 887
Query: 469 FAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLF 528
FAAA+ WVMNVVP STL ++++RGLIG HDWCE FSTYPRTYDL+HA +F
Sbjct: 888 FAAALKDKDVWVMNVVPPYG-PSTLRIVFDRGLIGTTHDWCEAFSTYPRTYDLLHAWTVF 1064
Query: 529 SLYQDK-CNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRW------NMKLVD 581
S Q K C+ ED++LEMDR++RP G +IIRD+ V+ VKK + + W +
Sbjct: 1065SDVQAKECSQEDLILEMDRLVRPTGFIIIRDKQSVIDFVKKYLTALHWEEVATADSSSDS 1244
Query: 582 HEDGPLVPEKVLIAVKQYWVT 602
+DG E + + K+ W+T
Sbjct: 1245DQDG---NEIIFVIQKKLWLT 1298
>TC81615 similar to GP|11994314|dbj|BAB02273. ankyrin-like protein
{Arabidopsis thaliana}, partial (69%)
Length = 1489
Score = 214 bits (544), Expect(3) = 5e-97
Identities = 107/241 (44%), Positives = 145/241 (59%), Gaps = 7/241 (2%)
Frame = +3
Query: 151 ANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIPI------NDG 204
AN P+ L EK+ QNW+ +G FPGGGT F +GADKYI +A+++ N G
Sbjct: 9 ANIPHTHLATEKSDQNWMVVKGEKIVFPGGGTHFHRGADKYIASIANMLNFPNNIINNGG 188
Query: 205 TVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLP 264
+R D GCGVAS+G YL S +V+AMS AP D HE Q+QFALERG+PA +GV GT +LP
Sbjct: 189 RLRNVFDVGCGVASFGGYLLSSDVLAMSLAPNDVHENQIQFALERGIPAYLGVLGTFRLP 368
Query: 265 YPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKE 324
YPSR+F++AHCSRC I W DG+ ++E+DR+LRPGGY+V S P E
Sbjct: 369 YPSRSFELAHCSRCRIDWLQRDGILLLELDRILRPGGYFVYSSPEA---------YAQDE 521
Query: 325 ELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVEFCES-SDPDDV 383
E R++ + ++CW+ ++K + IW K + R+ + C S DPD V
Sbjct: 522 EDRRIWREMSALVGRMCWKVAAKKNQTVIWVKPLTNDCYLKREPRTQPPLCSSDDDPDTV 701
Query: 384 W 384
W
Sbjct: 702 W 704
Score = 118 bits (296), Expect(3) = 5e-97
Identities = 59/128 (46%), Positives = 86/128 (67%), Gaps = 4/128 (3%)
Frame = +2
Query: 479 WVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFS-LYQDKCNT 537
WVMNVVP +TL +IY+RGL+G H+WCE FSTYPRTYDL+HA +FS + + +C+
Sbjct: 995 WVMNVVPENGP-NTLKIIYDRGLLGTVHNWCEAFSTYPRTYDLLHAWTIFSDIIERECSV 1171
Query: 538 EDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWN---MKLVDHEDGPLVPEKVLI 594
+DIL+EMDRILRP+ VII D+ V++ ++K + + W+ V+ + + VLI
Sbjct: 1172 QDILIEMDRILRPKAFVIIHDKRSVVMSIRKFLPALHWDAVTKSNVEQDSDTGEDDAVLI 1351
Query: 595 AVKQYWVT 602
K+ W+T
Sbjct: 1352 IQKKMWLT 1375
Score = 62.8 bits (151), Expect(3) = 5e-97
Identities = 36/92 (39%), Positives = 53/92 (57%), Gaps = 5/92 (5%)
Frame = +1
Query: 387 KLKACVT----PTPKVSGGDLKPFPDRLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVN 442
K+KAC+T K G DL P+P RL PPR++ + S+E + D +W++ V+
Sbjct: 712 KMKACITRYSDQVHKARGSDLSPWPARLTTPPPRLADFNY---SNEMFVKDMDVWQRRVS 882
Query: 443 AY-KKINSLLDSGRYRNIMDMNAGLGSFAAAI 473
Y K + + + RN+MDM A LGSFAAA+
Sbjct: 883 NYWKLLGNKIKPDTIRNVMDMKANLGSFAAAL 978
Score = 49.3 bits (116), Expect = 4e-06
Identities = 43/166 (25%), Positives = 76/166 (44%), Gaps = 11/166 (6%)
Frame = +3
Query: 423 PGVSSETYQNDNKMWKKHVNAYKKINSLLDSG-RYRNIMDMNAGLGSFAAAIHSSKSWVM 481
PG + ++ +K N N+++++G R RN+ D+ G+ SF + SS M
Sbjct: 90 PGGGTHFHRGADKYIASIANMLNFPNNIINNGGRLRNVFDVGCGVASFGGYLLSSDVLAM 269
Query: 482 NVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYP-RTYDLIHANGLFSLYQDKCNTEDI 540
++ P ++ + ERG+ YP R+++L H + + + +
Sbjct: 270 SLAPNDVHENQIQFALERGIPAYLGVLGTFRLPYPSRSFELAHCSRCRIDWLQRDGI--L 443
Query: 541 LLEMDRILRPEGAVI-------IRDEVDVLI--KVKKLIGGMRWNM 577
LLE+DRILRP G + +DE D I ++ L+G M W +
Sbjct: 444 LLELDRILRPGGYFVYSSPEAYAQDEEDRRIWREMSALVGRMCWKV 581
>AW776066 similar to GP|6752888|gb| unknown {Malus x domestica}, partial
(31%)
Length = 583
Score = 327 bits (839), Expect = 6e-90
Identities = 148/188 (78%), Positives = 167/188 (88%)
Frame = +1
Query: 176 RFPGGGTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAP 235
RFPGGGTQFPQGAD YI+QLASVIP+++G +RTALDTGCGVASWGAYL+ +NV+AMS AP
Sbjct: 1 RFPGGGTQFPQGADAYINQLASVIPLDNGMIRTALDTGCGVASWGAYLFKKNVIAMSIAP 180
Query: 236 RDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDR 295
RDSHEAQVQFALERGVPA+IGV GTI LP+PS AFDMAHCSRCLIPWGAN G+YM EVDR
Sbjct: 181 RDSHEAQVQFALERGVPAIIGVLGTIMLPFPSGAFDMAHCSRCLIPWGANGGLYMKEVDR 360
Query: 296 VLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQ 355
VLRPGGYW+LSGPPINWK N++ WQRP+ ELEEEQR+IE AK LCWEKK EK EIAIW+
Sbjct: 361 VLRPGGYWILSGPPINWKNNFRAWQRPENELEEEQRQIENTAKLLCWEKKHEKGEIAIWR 540
Query: 356 KMTDTESC 363
K + + C
Sbjct: 541 KALNIDEC 564
Score = 35.0 bits (79), Expect = 0.084
Identities = 27/107 (25%), Positives = 47/107 (43%), Gaps = 1/107 (0%)
Frame = +1
Query: 451 LDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCE 510
LD+G R +D G+ S+ A + M++ P + ++ + ERG+ I
Sbjct: 76 LDNGMIRTALDTGCGVASWGAYLFKKNVIAMSIAPRDSHEAQVQFALERGVPAIIGVLGT 255
Query: 511 GFSTYPR-TYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVII 556
+P +D+ H + L N + E+DR+LRP G I+
Sbjct: 256 IMLPFPSGAFDMAHCSRC--LIPWGANGGLYMKEVDRVLRPGGYWIL 390
>TC80791 similar to GP|16648931|gb|AAL24317.1 Unknown protein {Arabidopsis
thaliana}, partial (72%)
Length = 1890
Score = 202 bits (515), Expect(3) = 2e-83
Identities = 114/299 (38%), Positives = 160/299 (53%), Gaps = 6/299 (2%)
Frame = +2
Query: 71 VSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLHCMIP 130
V I+E N K + C A D+ PC+D RR RE YRERHCP EE C+IP
Sbjct: 470 VVAIEEGMLNGKSIEACPASEVDHMPCEDPRRNSQLSREMNYYRERHCPLPEETAVCLIP 649
Query: 131 APKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADK 190
P GY P WP+S + ++N P+ + K Q + + P + +
Sbjct: 650 PPNGYRVPVRWPESMHKIWHSNMPHNKIADRKGHQRVDETRRSTLYIPRWWHDVSRWSRT 829
Query: 191 YIDQLASVIPINDGTVRTALD-TGCGVASWGAYLWS-----RNVVAMSFAPRDSHEAQVQ 244
+ S I+ R + D T G+ WG W ++++ MSFAPRDSH++Q+Q
Sbjct: 830 IYQKT*S---IHSHKWRCSEDCT*HGM--WGC*FWRIFT*PQDILTMSFAPRDSHKSQIQ 994
Query: 245 FALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWV 304
FALERG+PA + + GT +LP+P+ FD+ HCSRCLIP+ A + Y +EVDR+LRPGGY V
Sbjct: 995 FALERGIPAFVAMLGTRRLPFPAFGFDLVHCSRCLIPFTAYNATYFIEVDRLLRPGGYLV 1174
Query: 305 LSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESC 363
+SGPP+ W K W ++ VAK LC+E+ + AIW+K +SC
Sbjct: 1175ISGPPVRWAKQEKEWS-----------DLQAVAKALCYEQITVHENTAIWKKPA-ADSC 1315
Score = 122 bits (305), Expect(3) = 2e-83
Identities = 68/153 (44%), Positives = 90/153 (58%), Gaps = 7/153 (4%)
Frame = +1
Query: 401 GDLKPFPDRLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKK-INSLLDSGRYRNI 459
G + +P RL A P R + + Y+ D K+W + V YK +N L + RN+
Sbjct: 1444 GTIPKWPKRLTAAPSR---SPLLKTGVDVYEADTKLWVQRVAHYKNSLNIKLGTPSIRNV 1614
Query: 460 MDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTY 519
MDMNA G FAAA+ WVMNVVP + TL I++RGLIG+YHDWCE FSTYP TY
Sbjct: 1615 MDMNALYGGFAAALKFDPVWVMNVVPA-QKPPTLDAIFDRGLIGVYHDWCEPFSTYPSTY 1791
Query: 520 DLIHANGLFSLYQDKCNTE------DILLEMDR 546
DLIHA + SL +D + D+++E+DR
Sbjct: 1792 DLIHAVSIESLIKDPATGKNQMYIGDLMVEIDR 1890
Score = 25.0 bits (53), Expect(3) = 2e-83
Identities = 11/22 (50%), Positives = 13/22 (59%), Gaps = 1/22 (4%)
Frame = +3
Query: 372 VEFCESS-DPDDVWYKKLKACV 392
+E C+ S D WY KLK CV
Sbjct: 1341 LELCDDSGDLSQAWYFKLKKCV 1406
>BG453377 similar to GP|6752888|gb| unknown {Malus x domestica}, partial
(29%)
Length = 534
Score = 303 bits (776), Expect = 1e-82
Identities = 140/179 (78%), Positives = 159/179 (88%)
Frame = +1
Query: 309 PINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQD 368
PI+WK NYK WQRPKE+LEEEQRKIE+VAK LCWEKKSEK EIA+WQK D+E+CR RQ+
Sbjct: 1 PIHWKANYKAWQRPKEDLEEEQRKIEDVAKLLCWEKKSEKNEIAVWQKTVDSETCRRRQE 180
Query: 369 DSSVEFCESSDPDDVWYKKLKACVTPTPKVSGGDLKPFPDRLYAIPPRVSSGSIPGVSSE 428
DS V+FCES+D +DVWYKK++ACVTP KV G DLKPFP RLYA+PP+++SGS+PGVS+E
Sbjct: 181 DSGVKFCESTDANDVWYKKMEACVTPNRKVHG-DLKPFPQRLYAVPPKIASGSVPGVSAE 357
Query: 429 TYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTI 487
TYQ+DNK WKKHVNAYKKIN LL SGRYRNIMDMNAGLGSFAAAI S K WVMNVVPTI
Sbjct: 358 TYQDDNKRWKKHVNAYKKINKLLGSGRYRNIMDMNAGLGSFAAAIQSPKLWVMNVVPTI 534
>AJ498686 similar to GP|6752888|gb| unknown {Malus x domestica}, partial
(27%)
Length = 750
Score = 296 bits (757), Expect = 2e-80
Identities = 134/177 (75%), Positives = 158/177 (88%)
Frame = +2
Query: 433 DNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKST 492
D+++WKKHVNAYK++N ++DSGRYRN+MDMNAG GSFAAA+ S K WVMNVVPTIAEK+T
Sbjct: 17 DSRLWKKHVNAYKRVNKIIDSGRYRNVMDMNAGFGSFAAALDSPKLWVMNVVPTIAEKAT 196
Query: 493 LGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEG 552
LGV++ERGLIGIYHDWCE FSTYPRTYDLIHANG+F+LY++ CN EDILLEMDRILRPEG
Sbjct: 197 LGVVFERGLIGIYHDWCEAFSTYPRTYDLIHANGVFTLYKNACNAEDILLEMDRILRPEG 376
Query: 553 AVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYWVTDGNSTST 609
AVI RD+V VL +VK++ GMRWN K+VDHEDGPL+ EKVL AVK+YWV N+TST
Sbjct: 377 AVIFRDQVGVLKQVKRIAKGMRWNTKMVDHEDGPLISEKVLYAVKRYWVAGDNTTST 547
Score = 41.2 bits (95), Expect = 0.001
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 2/103 (1%)
Frame = +2
Query: 201 INDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGT 260
I+ G R +D G S+ A L S + M+ P + +A + ERG+ + +
Sbjct: 71 IDSGRYRNVMDMNAGFGSFAAALDSPKLWVMNVVPTIAEKATLGVVFERGLIGIYHDWCE 250
Query: 261 IKLPYPSRAFDMAHCSRCLIPW--GANDGMYMMEVDRVLRPGG 301
YP R +D+ H + + N ++E+DR+LRP G
Sbjct: 251 AFSTYP-RTYDLIHANGVFTLYKNACNAEDILLEMDRILRPEG 376
>TC77957 similar to GP|15450900|gb|AAK96721.1 Unknown protein {Arabidopsis
thaliana}, partial (61%)
Length = 1689
Score = 223 bits (568), Expect(2) = 2e-76
Identities = 109/221 (49%), Positives = 142/221 (63%), Gaps = 11/221 (4%)
Frame = +1
Query: 73 QIDES-NSNTKVFKPCEARYTDYTPCQDQ----RRAMTFPRENMNYRERHCPPEEEKLHC 127
Q+DE N K F C+ R+++ PC D+ + M M + ERHCPP E + +C
Sbjct: 583 QVDEDDNIVPKSFPVCDDRHSELIPCLDRHLIYQLRMKLDLSVMEHYERHCPPAERRYNC 762
Query: 128 MIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQG 187
+IP P GY P WPKSRD V AN P+ L EK+ QNW+ +G FPGGGT F G
Sbjct: 763 LIPPPAGYKVPVKWPKSRDEVWKANIPHTHLAHEKSDQNWMVEKGEKIAFPGGGTHFHYG 942
Query: 188 ADKYIDQLASVIPI------NDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEA 241
ADKYI +A+++ N+G +RT LD GCGVAS+G YL S +++ MS AP D H+
Sbjct: 943 ADKYIASMANMLNFSNNNLNNEGRLRTVLDVGCGVASFGGYLLSSDIITMSLAPNDVHQN 1122
Query: 242 QVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPW 282
Q+QFALERG+PA +GV GT +LPYPSR+F++AHCSRC I W
Sbjct: 1123QIQFALERGIPAYLGVLGTKRLPYPSRSFELAHCSRCRIDW 1245
Score = 81.6 bits (200), Expect(2) = 2e-76
Identities = 45/142 (31%), Positives = 78/142 (54%), Gaps = 5/142 (3%)
Frame = +2
Query: 286 DGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKK 345
DG+ ++E+DRVLRPGGY+ S P + + + +E L +++ ++ ++CW
Sbjct: 1256 DGILLLELDRVLRPGGYFAYSSP--------EAYAQDEENL-RIWKEMSDLVGRMCWRIA 1408
Query: 346 SEKAEIAIWQKMTDTESCRSRQDDSSVEFCES-SDPDDVWYKKLKACVTP----TPKVSG 400
S+K + IWQK + + R+ + C+S +DPD V+ ++ C+TP K G
Sbjct: 1409 SKKEQTVIWQKPLTNDCYKKREPGTRPPLCQSDADPDAVFGVNMEVCITPYSEHDNKAKG 1588
Query: 401 GDLKPFPDRLYAIPPRVSSGSI 422
L P+P RL + PPR++ +I
Sbjct: 1589 SGLAPWPARLTSPPPRLADLAI 1654
>TC90244 similar to GP|15450749|gb|AAK96646.1 At2g39750/T5I7.5 {Arabidopsis
thaliana}, partial (49%)
Length = 1319
Score = 213 bits (541), Expect(3) = 6e-76
Identities = 121/295 (41%), Positives = 174/295 (58%), Gaps = 13/295 (4%)
Frame = +3
Query: 184 FPQGADKYIDQLASVIP-INDGTVRTA--LDTGCGVASWGAYLWSRNVVAMSFAPRDSHE 240
F GAD+Y+D ++ +IP I G T+ L GCGVAS+GAYL RNV+ MS AP+D HE
Sbjct: 477 FIHGADEYLDHISKMIPEITFGRHITSCSLMLGCGVASFGAYLLQRNVITMSVAPKDVHE 656
Query: 241 AQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPG 300
Q+QFALERGVPA++ F T +L YPS+AFD+ HCSRC I W +DG+ ++EV+R+LR G
Sbjct: 657 NQIQFALERGVPAMVAAFATRRLLYPSQAFDLIHCSRCRINWTRDDGILLLEVNRMLRAG 836
Query: 301 GYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDT 360
GY+V + P+ + +E LEE+ ++ + +LCW+ + IA+WQK D
Sbjct: 837 GYFVWAAQPV---------YKHEEALEEQWEEMLNLTTRLCWKFLKKDGYIAVWQKPFDN 989
Query: 361 ESCRSRQDDSSVEFCE-SSDPDDVWYKKLKACVTPTPKVS-GGDLKPFPDRLYAIPPRVS 418
+R+ + C+ S DPD+VWY LKAC++ PK ++ +P RL P R+
Sbjct: 990 SCYLNREAGTKPPLCDPSDDPDNVWYVDLKACISELPKNEYEANITDWPARLQTPPNRLQ 1169
Query: 419 SGSIPGVSS--ETYQNDNKMWKKHVNAY------KKINSLLDSGRYRNIMDMNAG 465
S + S E ++ ++K W + + AY KKI R RN+MDM G
Sbjct: 1170SIKVDAFISRKELFKAESKYWNEIIEAYVRALHWKKI-------RLRNVMDMRXG 1313
Score = 85.9 bits (211), Expect(3) = 6e-76
Identities = 41/89 (46%), Positives = 53/89 (59%), Gaps = 2/89 (2%)
Frame = +1
Query: 82 KVFKPCEARYTDYTPCQDQRRAMT-FPRENMNYR-ERHCPPEEEKLHCMIPAPKGYVTPF 139
K F C ++Y PC D A+ P R ERHCP + +KL+C++PAPKGY P
Sbjct: 163 KKFGLCSRGMSEYIPCLDNVEAIKKLPSTEKGERFERHCPEDGKKLNCLVPAPKGYRAPI 342
Query: 140 PWPKSRDYVPYANAPYKSLTVEKAIQNWI 168
PWPKSRD V ++N P+ L +K QNWI
Sbjct: 343 PWPKSRDEVWFSNVPHTRLVEDKGGQNWI 429
Score = 30.0 bits (66), Expect = 2.7
Identities = 26/98 (26%), Positives = 44/98 (44%), Gaps = 7/98 (7%)
Frame = +3
Query: 462 MNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYP-RTYD 520
+ G+ SF A + M+V P ++ + ERG+ + + YP + +D
Sbjct: 570 LGCGVASFGAYLLQRNVITMSVAPKDVHENQIQFALERGVPAMVAAFATRRLLYPSQAFD 749
Query: 521 LIHANGLFSLYQDKCN---TED---ILLEMDRILRPEG 552
LIH + +C T D +LLE++R+LR G
Sbjct: 750 LIHCS--------RCRINWTRDDGILLLEVNRMLRAGG 839
Score = 25.8 bits (55), Expect(3) = 6e-76
Identities = 9/10 (90%), Positives = 10/10 (100%)
Frame = +2
Query: 175 FRFPGGGTQF 184
F+FPGGGTQF
Sbjct: 449 FKFPGGGTQF 478
>TC91665 similar to PIR|T02472|T02472 hypothetical protein At2g45750
[imported] - Arabidopsis thaliana, partial (35%)
Length = 991
Score = 280 bits (715), Expect = 1e-75
Identities = 135/255 (52%), Positives = 183/255 (70%), Gaps = 9/255 (3%)
Frame = +1
Query: 355 QKMTDTESCR-SRQDDSSVEFCESS-DPDDVWYKKLKACVTPTP------KVSGGDLKPF 406
QK T+ C+ R+ + FCE + DPD WY KL C+TP P +V+GG++ +
Sbjct: 1 QKPTNHMHCKVKRKIFKTRPFCEEAQDPDMAWYTKLDTCLTPLPDVNNVKEVAGGEISKW 180
Query: 407 PDRLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINS-LLDSGRYRNIMDMNAG 465
P RL +IPPR+S S+ G++ E ++ + ++WKK + YK ++S L ++GRYRN++DMN+
Sbjct: 181 PKRLTSIPPRISGESLKGITPEMFKENTELWKKRESYYKTLDSQLAETGRYRNLLDMNSY 360
Query: 466 LGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHAN 525
LG FAAA+ WVMN+VP AE +TLGVIYERGLIG Y +WCE STYPRTYD IH +
Sbjct: 361 LGGFAAALVDDPVWVMNIVPVEAEINTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHGD 540
Query: 526 GLFSLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDG 585
+FSLYQ +C+ E+ILLEMDRILRP+G+VI+RD VDVL KVK + M+W+ K+ DHE+G
Sbjct: 541 SVFSLYQHRCSMENILLEMDRILRPQGSVILRDNVDVLTKVKSIADEMQWDAKIRDHEEG 720
Query: 586 PLVPEKVLIAVKQYW 600
P EK+L+AVKQYW
Sbjct: 721 PYQKEKILVAVKQYW 765
Score = 30.4 bits (67), Expect = 2.1
Identities = 45/220 (20%), Positives = 77/220 (34%), Gaps = 4/220 (1%)
Frame = +1
Query: 141 WPKSRDYVP--YANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASV 198
WPK +P + K +T E +N T+ + + Y L S
Sbjct: 178 WPKRLTSIPPRISGESLKGITPEMFKEN---------------TELWKKRESYYKTLDSQ 312
Query: 199 IPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVF 258
+ G R LD + + A L V M+ P ++ + ERG+ +
Sbjct: 313 LA-ETGRYRNLLDMNSYLGGFAAALVDDPVWVMNIVPVEAEINTLGVIYERGLIGTYQNW 489
Query: 259 GTIKLPYPSRAFDMAHCSRCLIPWGANDGM--YMMEVDRVLRPGGYWVLSGPPINWKVNY 316
YP R +D H + M ++E+DR+LRP G +L
Sbjct: 490 CEAMSTYP-RTYDFIHGDSVFSLYQHRCSMENILLEMDRILRPQGSVIL----------- 633
Query: 317 KPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQK 356
+ + K++ +A ++ W+ K E +QK
Sbjct: 634 -------RDNVDVLTKVKSIADEMQWDAKIRDHEEGPYQK 732
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 188 bits (478), Expect(3) = 2e-69
Identities = 104/252 (41%), Positives = 147/252 (58%), Gaps = 9/252 (3%)
Frame = +1
Query: 172 GNVFRFPGGGTQFPQGADKYIDQLASVIP-INDGT-VRTALDTGCGVASWGAYLWSRNVV 229
G FPGGGTQF GA YID + +P I G R LD GCGVAS+G +L+ R+V+
Sbjct: 1342 GEYLTFPGGGTQFKHGALHYIDFIQETLPDIAWGKRTRVILDVGCGVASFGGFLFDRDVL 1521
Query: 230 AMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMY 289
AMS AP+D HEAQVQFALERG+PA+ V GT +LP+P R FD HC+RC +PW G
Sbjct: 1522 AMSLAPKDEHEAQVQFALERGIPAISAVMGTKRLPFPGRVFDAVHCARCRVPWHIEGGKL 1701
Query: 290 MMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWE------ 343
++E++RVLRPGG++V S PI +Q+ E++ E +++ + K +CWE
Sbjct: 1702 LLELNRVLRPGGFFVWSATPI--------YQKLPEDV-EIWNEMKALTKSICWELVSISK 1854
Query: 344 KKSEKAEIAIWQKMTDTESCRSRQDDSSVEFCESSDPDD-VWYKKLKACVTPTPKVSGGD 402
K +AI++K + C ++ + C+ SD + WY KL+AC+ P S
Sbjct: 1855 DKVNGVGVAIYKKPLSND-CYEQRSKNEPPMCQKSDDHNAAWYIKLQACIHKVPVSSSER 2031
Query: 403 LKPFPDRLYAIP 414
+P++ A P
Sbjct: 2032 GSQWPEKWPARP 2067
Score = 57.0 bits (136), Expect(3) = 2e-69
Identities = 34/74 (45%), Positives = 44/74 (58%), Gaps = 1/74 (1%)
Frame = +2
Query: 428 ETYQNDNKMWKKHVNAYKKINSL-LDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPT 486
E + DNK WK+ V+ +N L + RN+MDMN+ G FAAA+ WVMNVV +
Sbjct: 2129 EDFAADNKHWKRVVSK-SYLNGLGIQWSNVRNVMDMNSIYGGFAAALKELNIWVMNVV-S 2302
Query: 487 IAEKSTLGVIYERG 500
I TL +IYERG
Sbjct: 2303 IDSADTLPIIYERG 2344
Score = 57.0 bits (136), Expect(3) = 2e-69
Identities = 31/86 (36%), Positives = 44/86 (51%), Gaps = 3/86 (3%)
Frame = +2
Query: 73 QIDESNSNTKVFKPCEARY-TDYTPCQDQRRAMTFPRENMNY--RERHCPPEEEKLHCMI 129
+I ES+ +K C D+ PC D + + R +Y RERHCP EE C++
Sbjct: 1040 EIQESSKTGYNWKVCNVTAGPDFIPCLDNWKVIRSLRSTKHYEHRERHCP--EEPPTCLV 1213
Query: 130 PAPKGYVTPFPWPKSRDYVPYANAPY 155
P+GY WPKSR+ + Y N P+
Sbjct: 1214 SLPEGYKCSIEWPKSREKIWYYNVPH 1291
Score = 34.7 bits (78), Expect = 0.11
Identities = 30/139 (21%), Positives = 58/139 (41%), Gaps = 16/139 (11%)
Frame = +1
Query: 455 RYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFST 514
R R I+D+ G+ SF + M++ P ++ + ERG+ I
Sbjct: 1447 RTRVILDVGCGVASFGGFLFDRDVLAMSLAPKDEHEAQVQFALERGIPAISAVMGTKRLP 1626
Query: 515 YP-RTYDLIHANGLFSLYQDKCNTE------DILLEMDRILRPEGAVI---------IRD 558
+P R +D +H +C +LLE++R+LRP G + + +
Sbjct: 1627 FPGRVFDAVHCA--------RCRVPWHIEGGKLLLELNRVLRPGGFFVWSATPIYQKLPE 1782
Query: 559 EVDVLIKVKKLIGGMRWNM 577
+V++ ++K L + W +
Sbjct: 1783 DVEIWNEMKALTKSICWEL 1839
>TC81625 similar to PIR|T02472|T02472 hypothetical protein At2g45750
[imported] - Arabidopsis thaliana, partial (45%)
Length = 1431
Score = 250 bits (638), Expect = 1e-66
Identities = 117/212 (55%), Positives = 150/212 (70%), Gaps = 7/212 (3%)
Frame = +2
Query: 247 LERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLS 306
L+ GVPAVIGVF + +LP+PSRAFDMAHCSRCLIPW DG+Y+ EVDR+LRPGGYW+LS
Sbjct: 803 LKEGVPAVIGVFASKRLPFPSRAFDMAHCSRCLIPWPEYDGLYLNEVDRILRPGGYWILS 982
Query: 307 GPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSR 366
GPPI+WK +K W+R KE+L EEQ KIE VAK LCW K EK +IAIWQK + C+
Sbjct: 983 GPPIHWKRYWKGWERTKEDLNEEQTKIENVAKSLCWNKIVEKGDIAIWQKPKNHLDCKFT 1162
Query: 367 QDDSSVEFC-ESSDPDDVWYKKLKACVTPTPKV------SGGDLKPFPDRLYAIPPRVSS 419
Q+ FC E ++PD WY ++ C+ P PKV SGG+L +P RL + PPR+S
Sbjct: 1163 QNR---PFCQEQNNPDKAWYTNMQTCLNPLPKVSNKEEISGGELNNWPQRLKSTPPRISK 1333
Query: 420 GSIPGVSSETYQNDNKMWKKHVNAYKKINSLL 451
G+I V+ T+ DN++W K V+ YKK+N+ L
Sbjct: 1334 GTIKSVTPPTFSKDNQLWNKRVSYYKKVNNQL 1429
Score = 173 bits (438), Expect(2) = 1e-62
Identities = 88/201 (43%), Positives = 116/201 (56%), Gaps = 11/201 (5%)
Frame = +3
Query: 9 NRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAEC-----------DV 57
N ++++ FI + LC Y LG++Q + S ++ K +C
Sbjct: 63 NCKKTNLYSFIAL-LCIISYFLGSYQNNT-STSTSKNIQNKKVTPKCLQKNPTNTKTKTK 236
Query: 58 VPNLSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERH 117
NL F SHH + S + + + PC TDYTPC+D R++ + R+ M YRERH
Sbjct: 237 TTNLDFFSHH-NLTNPTTTSTTKSSQYPPCTPSLTDYTPCEDHTRSLKYTRDKMIYRERH 413
Query: 118 CPPEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRF 177
CP + E L C +PAP GY PFPWP SRD YAN PY+ LTVEKA QNWI+++G+ FRF
Sbjct: 414 CPKKHEILKCRVPAPNGYKNPFPWPTSRDMAWYANVPYRHLTVEKAGQNWIRFDGDKFRF 593
Query: 178 PGGGTQFPQGADKYIDQLASV 198
PGGGT FP GADKYID + V
Sbjct: 594 PGGGTMFPNGADKYIDDIGKV 656
Score = 85.9 bits (211), Expect(2) = 1e-62
Identities = 40/75 (53%), Positives = 55/75 (73%)
Frame = +1
Query: 195 LASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAV 254
L +I + DG+VRTA+DTGCGVASWGAYL SR+++ +S AP+D+HEAQVQFALER
Sbjct: 646 LGKLIDLEDGSVRTAVDTGCGVASWGAYLLSRDILTVSIAPKDTHEAQVQFALERRCSCS 825
Query: 255 IGVFGTIKLPYPSRA 269
F ++ +P ++
Sbjct: 826 YWCFCFKEITFPFKS 870
>TC79561 similar to GP|14194107|gb|AAK56248.1 At1g77260/T14N5_19
{Arabidopsis thaliana}, partial (33%)
Length = 1092
Score = 239 bits (611), Expect = 2e-63
Identities = 122/266 (45%), Positives = 172/266 (63%), Gaps = 6/266 (2%)
Frame = +2
Query: 51 NNAECDVVPNLSFD-SHHAGEVSQIDESNSNTKVFKPC-EARYTDYTPCQD--QRRAMTF 106
NN + + + N S + +++ + + + E K +K C + R DY PC D + A
Sbjct: 293 NNDDDETLLNWSLNITNNNSDTNFVVEEVKKVKKYKICKDVRMVDYIPCLDNFEEIAKFN 472
Query: 107 PRENMNYRERHCPPEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQN 166
E ERHCP +E+ L+C++P PKGY P WPKSRD + ++N P+ L +K QN
Sbjct: 473 GSERGEKYERHCPEQEKGLNCVVPRPKGYRKPILWPKSRDEIWFSNVPHTRLVEDKGGQN 652
Query: 167 WIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIP-INDG-TVRTALDTGCGVASWGAYLW 224
WI + + F FPGGGTQF GADKY+DQ++ ++P I G R ALD GCGVAS+GA+L
Sbjct: 653 WISRKKDKFVFPGGGTQFIHGADKYLDQISEMVPDIAFGYNTRVALDIGCGVASFGAFLM 832
Query: 225 SRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGA 284
RNV +S AP+D HE Q+QFALERGVPA++ VF T +L +PS+AFD+ HCSRC I W
Sbjct: 833 QRNVTTLSIAPKDVHENQIQFALERGVPALVAVFATHRLLFPSQAFDLIHCSRCRINWTR 1012
Query: 285 NDGMYMMEVDRVLRPGGYWVLSGPPI 310
+DG+ ++E +R+LR G Y+V + P+
Sbjct: 1013DDGILLLEANRLLRAGRYFVWAAQPV 1090
>TC82332 similar to GP|16604525|gb|AAL24268.1 AT4g00750/F15P23_1
{Arabidopsis thaliana}, partial (95%)
Length = 658
Score = 209 bits (532), Expect = 2e-54
Identities = 96/148 (64%), Positives = 118/148 (78%)
Frame = +2
Query: 462 MNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDL 521
MNA LG FAA++ WVMNVVP A+ TLG IYERGLIG+YH+WCE STYPRTYDL
Sbjct: 2 MNAYLGGFAASLVKYPIWVMNVVPIQAKVDTLGAIYERGLIGMYHNWCEAMSTYPRTYDL 181
Query: 522 IHANGLFSLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVD 581
IHA+ +FSLY D+C EDILLEMDRILRPEG+VIIRD+VD+L+KVK +I G+ W ++VD
Sbjct: 182 IHADSVFSLYNDRCELEDILLEMDRILRPEGSVIIRDDVDILVKVKSIINGLEWESQIVD 361
Query: 582 HEDGPLVPEKVLIAVKQYWVTDGNSTST 609
HEDGPL EK+L AVK+YW + ++ +T
Sbjct: 362 HEDGPLEREKLLFAVKKYWTSPASNDNT 445
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,526,573
Number of Sequences: 36976
Number of extensions: 327367
Number of successful extensions: 1861
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 1738
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1783
length of query: 610
length of database: 9,014,727
effective HSP length: 102
effective length of query: 508
effective length of database: 5,243,175
effective search space: 2663532900
effective search space used: 2663532900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146568.1


