

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
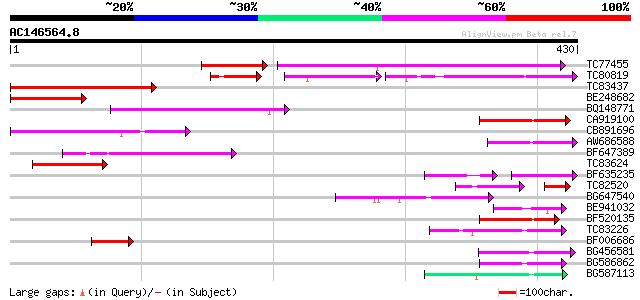
Query= AC146564.8 - phase: 0 /pseudo
(430 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 184 3e-61
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 74 8e-29
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 96 2e-20
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 91 7e-19
BQ148771 66 3e-11
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 65 4e-11
CB891696 65 5e-11
AW686588 59 3e-09
BF647389 57 1e-08
TC83624 homologue to PIR|G84581|G84581 copia-like retroelement p... 55 5e-08
BF635235 41 2e-07
TC82520 38 9e-07
BG647540 49 5e-06
BE941032 48 8e-06
BF520135 48 8e-06
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 47 1e-05
BF006686 47 2e-05
BG456581 45 4e-05
BG586862 45 7e-05
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 44 9e-05
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 184 bits (466), Expect(2) = 3e-61
Identities = 100/223 (44%), Positives = 130/223 (57%), Gaps = 5/223 (2%)
Frame = -2
Query: 204 SWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNGETTRFWLDHWVGNEALYLTFPRLF 263
+W + ASRWW DLMSLE V WF RE++RKVG+G ++ FW D W + L +FPR F
Sbjct: 796 NWPAYASRWWKDLMSLEEVGRVRWFPRELIRKVGDGRSSFFWKDAWDSSVPLRESFPRAF 617
Query: 264 SISSQKEAMVGEVW---VDG-DWNLTWRR-SLFVWEEGLIHILLDELEGREVSEPMDSWW 318
+ G++W +G W L WRR LF WE+ + LL LEG + D W
Sbjct: 616 FPYRLLKMGCGDLWDMNAEGVRWRLYWRRLELFEWEKERLLELLGRLEGVVLRYWADIWV 437
Query: 319 WKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGSVWKSPTPFKVVVFSWQSLLDRI 378
WK ++ G+ SV+S Y L L++ + L + ++F +WKS P KV+ FSW LDRI
Sbjct: 436 WKPDKEGVFSVNSCYFLLQNLRLLEDRLSYEEEVIFRELWKSKAPAKVLAFSWTLFLDRI 257
Query: 379 PTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFKV 421
PT NL KRR+L E S RCVFC ET HLFLHCD+ KV
Sbjct: 256 PTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHCDVISKV 128
Score = 69.7 bits (169), Expect(2) = 3e-61
Identities = 32/50 (64%), Positives = 40/50 (80%)
Frame = -3
Query: 146 CLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL*KDVLRCKYGSRIN 195
CLPR KGGLGVRD++LVN+SLLAKW WRL+Q + L K+VL YG R++
Sbjct: 969 CLPRCKGGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYGPRVS 820
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 73.6 bits (179), Expect(3) = 8e-29
Identities = 49/148 (33%), Positives = 72/148 (48%), Gaps = 3/148 (2%)
Frame = +2
Query: 286 WRRSLFVWEEGLIH---ILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMP 342
W R LFVWEE + ILL+ ++ D W W L+ + YS+ V +
Sbjct: 428 WTRRLFVWEEECVRECCILLNNFVLQD--NVNDKWRWLLDP------VNGYSVKVFYRYI 583
Query: 343 PEPLENTKAMVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCD 402
+ + VW P KV +F W+ L +R+PTKDNL+ R +L ++ CV
Sbjct: 584 TSTGHISDRSLVDDVWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVL-LATNAACVCGC 760
Query: 403 QVGETAAHLFLHCDMSFKVWSMVYCRLG 430
E+ HLFLHC++ +WS+V LG
Sbjct: 761 VDSESTTHLFLHCNVFCSLWSLVRNWLG 844
Score = 57.8 bits (138), Expect(3) = 8e-29
Identities = 28/77 (36%), Positives = 37/77 (47%), Gaps = 3/77 (3%)
Frame = +3
Query: 209 ASRWWLDLMSLEGVVG---ANWFNREMVRKVGNGETTRFWLDHWVGNEALYLTFPRLFSI 265
+S WW + + VG NWF + VG+G FW D W G+ L L +PRL +
Sbjct: 174 SSMWWKTICKVREGVGEGVGNWFEENIRMVVGDGRDAFFWYDTWAGDVPLRLKYPRLLDL 353
Query: 266 SSQKEAMVGEVWVDGDW 282
+ KE VG W G W
Sbjct: 354 AMDKECKVGLTW--GGW 398
Score = 33.9 bits (76), Expect(3) = 8e-29
Identities = 18/39 (46%), Positives = 24/39 (61%)
Frame = +1
Query: 153 GLGVRDMKLVNISLLAKWRWRLVQPEQPL*KDVLRCKYG 191
GLGV N+SLL KW WRL+ ++ L VL+ +YG
Sbjct: 31 GLGVGAF---NLSLLGKWCWRLLVDKEGLWHRVLKARYG 138
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 96.3 bits (238), Expect = 2e-20
Identities = 48/111 (43%), Positives = 72/111 (64%)
Frame = +2
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
L+A L F SGLKVNF KS L+ VN+ ++ A + ++ + G PF YLG+P+ N
Sbjct: 611 LRAALVIF*AMSGLKVNFHKSGLVCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNS 790
Query: 61 RRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKL 111
RRLS WEP+++ +K RL W ++++S GGR+VLL +VL ++ + L KL
Sbjct: 791 RRLSFWEPIVNRIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYALPSSKL 943
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 91.3 bits (225), Expect = 7e-19
Identities = 44/58 (75%), Positives = 50/58 (85%)
Frame = +3
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGA 58
LKALL+GFEMASGLKVNF KS LIG+NV R+FM+AAC F+NCRE S PF YLGLP G+
Sbjct: 228 LKALLQGFEMASGLKVNFHKSSLIGINVPRDFMEAACRFLNCREESIPFIYLGLPGGS 401
>BQ148771
Length = 680
Score = 65.9 bits (159), Expect = 3e-11
Identities = 39/141 (27%), Positives = 70/141 (48%), Gaps = 5/141 (3%)
Frame = -3
Query: 77 LNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVVRIQREFLWGGVNGGKK 136
L +W ++SL RV L +V+ A+P + + +P + + ++QR+F+WG ++
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 137 VCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL*KDVLRCKYGSRIN- 195
V W T+ P+ GLG+R + ++N + + K W + L +V+R KY +
Sbjct: 393 YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKYQRSESL 214
Query: 196 ---FLLDPIYNS-WSSLASRW 212
FL P +S W +L W
Sbjct: 213 EEIFLEKPTDSSLWKALVKLW 151
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 65.5 bits (158), Expect = 4e-11
Identities = 31/69 (44%), Positives = 43/69 (61%)
Frame = -2
Query: 357 VWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCD 416
+W P KV VF+W+ L DR+PTK NL+ R ++PTE+ + CV E+A HLFL C
Sbjct: 584 IWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGL-CVSGCGALESAQHLFLSCS 408
Query: 417 MSFKVWSMV 425
+WS+V
Sbjct: 407 YFASLWSLV 381
>CB891696
Length = 638
Score = 65.1 bits (157), Expect = 5e-11
Identities = 47/140 (33%), Positives = 73/140 (51%), Gaps = 3/140 (2%)
Frame = +1
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
+K ++ FE+AS L VNF KS LI +NV F ++ C+ F+YLG+ VG NP
Sbjct: 22 MKTIVSYFELASSLWVNFLKSGLINLNVIGHF*GW*NIYIKCKVH*VIFKYLGILVGENP 201
Query: 61 RRLSTWEPLLDVLKKRLNSWGNK---YVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWK 117
R++ E LL +L L SW N + G + + N Y + K+PVKV +
Sbjct: 202 CRVNM*ELLLKLLTN*LGSWWNTK*LWTQNGFSQIHAK*ISQ---NIYFSLMKIPVKV*E 372
Query: 118 RVVRIQREFLWGGVNGGKKV 137
+ +++ +FL G + KK+
Sbjct: 373 LISQLKTQFL*GNLKVTKKI 432
Score = 29.6 bits (65), Expect = 2.3
Identities = 14/43 (32%), Positives = 22/43 (50%)
Frame = +2
Query: 384 LLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFKVWSMVY 426
LL RI S CVFC E++ HLF C++ + + ++
Sbjct: 434 LLHCRIFQQVSRPTCVFCGCTNESS*HLFRICNLFLRFYYRIF 562
>AW686588
Length = 567
Score = 59.3 bits (142), Expect = 3e-09
Identities = 29/68 (42%), Positives = 41/68 (59%)
Frame = +1
Query: 363 PFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFKVW 422
P KV + +W+ + DR+PTK NL++RR L E++ CV + ETA HLFLHC VW
Sbjct: 145 PLKVSILAWRLIRDRLPTKANLVRRRCLAVEAA-GCVVGCGIAETANHLFLHCATFGAVW 321
Query: 423 SMVYCRLG 430
+ +G
Sbjct: 322 QHIRAWIG 345
>BF647389
Length = 404
Score = 57.4 bits (137), Expect = 1e-08
Identities = 38/132 (28%), Positives = 67/132 (49%)
Frame = +2
Query: 41 NCREGSPPFQYLGLPVGANPRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNA 100
N +G P +YLG+P+ + ++L + + ++ R+ + +K +S G + L+ VL
Sbjct: 14 NLIKGKLPSRYLGVPLSS--KKLYVIQRVKKIIC-RIEN*SSKLLSYAGSLQLIKIVLFG 184
Query: 101 IPNFYLAFFKLPVKVWKRVVRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMK 160
+ ++ F LP KV K + R FL G +G K + +CLP+ GG V D+K
Sbjct: 185 VQPYWSQVFVLP*KVIKLIQTTCRIFL*TGKSGTSKRALIAREHICLPKTAGGWNVIDLK 364
Query: 161 LVNISLLAKWRW 172
+ N + + K W
Sbjct: 365 VXNQTAICKLXW 400
>TC83624 homologue to PIR|G84581|G84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 831
Score = 55.1 bits (131), Expect = 5e-08
Identities = 24/57 (42%), Positives = 39/57 (68%)
Frame = +1
Query: 18 FAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANPRRLSTWEPLLDVLK 74
F++ + +N+ F++A+ NF+ C PF +LGLP+GANP+R ST +P+LD L+
Sbjct: 256 FSRVNFMALNLEESFVEASPNFLLCNVNEVPFCFLGLPIGANPKRSSTRKPVLDSLQ 426
>BF635235
Length = 402
Score = 41.2 bits (95), Expect(2) = 2e-07
Identities = 18/50 (36%), Positives = 27/50 (54%)
Frame = +3
Query: 381 KDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFKVWSMVYCRLG 430
+ NL +RR++ + CVF + E++ HLF+ CD F VW R G
Sbjct: 192 RQNLFRRRVIIDHRGINCVF*GFLVESSCHLFVTCDFVFHVWYSHASRFG 341
Score = 31.6 bits (70), Expect(2) = 2e-07
Identities = 20/56 (35%), Positives = 25/56 (43%)
Frame = +2
Query: 315 DSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGSVWKSPTPFKVVVFS 370
D W WK + G VSS Y++ L+ VWKS P KV+VFS
Sbjct: 32 DCWRWKHDSSGSFLVSSGYAVISDFSQVGPNLK---------VWKS*FPSKVIVFS 172
>TC82520
Length = 833
Score = 37.7 bits (86), Expect(2) = 9e-07
Identities = 18/52 (34%), Positives = 30/52 (57%)
Frame = +3
Query: 339 LQMPPEPLENTKAMVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRIL 390
L + +PL+ + VW+ P KV +F W+ +R+PTK NL++R +L
Sbjct: 141 LTISGQPLDRNQV---DDVWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHVL 287
Score = 32.7 bits (73), Expect(2) = 9e-07
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = +2
Query: 406 ETAAHLFLHCDMSFKVWSMV 425
ETA HLFLHCD+ +WS V
Sbjct: 329 ETATHLFLHCDIFGSLWSHV 388
>BG647540
Length = 625
Score = 48.5 bits (114), Expect = 5e-06
Identities = 41/131 (31%), Positives = 58/131 (43%), Gaps = 11/131 (8%)
Frame = +3
Query: 248 HWVGNEALYLTFPRLFSISSQKEAMVGE---VWV----DGDWNLTWRRSLFVWE----EG 296
H+V LTF ++F + E +WV DG W++ WRRSLF+WE G
Sbjct: 120 HFVYTFLFSLTFLKIFVDYLFLHYQLSEWNKLWVNNSTDGYWDVNWRRSLFLWEFNTCFG 299
Query: 297 LIHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGS 356
+ ++E +V LEE G SV+S Y ++ L + VF
Sbjct: 300 SCWRIWTDIEEGKVRMYGGG---DLEENGWFSVNSMYKKLEGMRFEEGSLTEMRVKVFTH 470
Query: 357 VWKSPTPFKVV 367
+WKS P KVV
Sbjct: 471 IWKSSAPSKVV 503
>BE941032
Length = 435
Score = 47.8 bits (112), Expect = 8e-06
Identities = 25/60 (41%), Positives = 34/60 (56%), Gaps = 5/60 (8%)
Frame = +2
Query: 368 VFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGE-----TAAHLFLHCDMSFKVW 422
+F W LL+R PTKDNLLKR + + ++ VGE A HLFLHC+ ++W
Sbjct: 149 IFLWCVLLNRFPTKDNLLKRGV------ISAIYQSCVGECGNLYDATHLFLHCNFFRQIW 310
>BF520135
Length = 202
Score = 47.8 bits (112), Expect = 8e-06
Identities = 23/61 (37%), Positives = 38/61 (61%)
Frame = +3
Query: 357 VWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCD 416
+W+ KV +F+W+ R+PTK N+ KR I+ ++ + CV ++ E+ HLFLHCD
Sbjct: 15 LWRKEDLSKVSIFAWRLFHGRLPTKANVFKRGIVHHDAHM-CVTRCRLIESDVHLFLHCD 191
Query: 417 M 417
+
Sbjct: 192 V 194
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 47.0 bits (110), Expect = 1e-05
Identities = 32/109 (29%), Positives = 49/109 (44%), Gaps = 5/109 (4%)
Frame = +3
Query: 319 WKLEEGGICSVSSSYSLHVKLQMPPEPLENT-----KAMVFGSVWKSPTPFKVVVFSWQS 373
W GI SV S Y+ Q + + NT + +++ +W T + V W+
Sbjct: 6 WMHNPTGIYSVKSGYNTLRTWQT--QQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRI 179
Query: 374 LLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFKVW 422
L D +P + +L KR I + C C ET HLF+ C +S +VW
Sbjct: 180 LNDSLPVRSSLRKRGI---QCYPLCPRCHSKTETITHLFMSCPLSKRVW 317
>BF006686
Length = 325
Score = 46.6 bits (109), Expect = 2e-05
Identities = 20/32 (62%), Positives = 24/32 (74%)
Frame = +3
Query: 63 LSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLL 94
L WEPLL+ + K L SWGNK +S GGR+VLL
Sbjct: 228 LPMWEPLLEHVNKMLKSWGNKLLSFGGRIVLL 323
>BG456581
Length = 683
Score = 45.4 bits (106), Expect = 4e-05
Identities = 25/74 (33%), Positives = 34/74 (45%)
Frame = +2
Query: 356 SVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHC 415
++W S P W+ DR+PT DNL R C FC + ET+ HLFL C
Sbjct: 77 TIWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCALVSM---CSFCLEQVETSDHLFLRC 247
Query: 416 DMSFKVWSMVYCRL 429
+WS + +L
Sbjct: 248 KFVVTLWSWLCSQL 289
>BG586862
Length = 804
Score = 44.7 bits (104), Expect = 7e-05
Identities = 25/66 (37%), Positives = 35/66 (52%)
Frame = -1
Query: 357 VWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCD 416
VW T + F W+ L + +P KD L KR I S+ C C+ ET HLFL+C+
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGI---RCSLLCPRCESKIETVQHLFLNCE 484
Query: 417 MSFKVW 422
++ K W
Sbjct: 483 VTQKEW 466
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 44.3 bits (103), Expect = 9e-05
Identities = 29/113 (25%), Positives = 45/113 (39%), Gaps = 4/113 (3%)
Frame = -3
Query: 315 DSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAM----VFGSVWKSPTPFKVVVFS 370
DS+ W+ + G SV S Y + + ++ VWK T KV F
Sbjct: 426 DSYSWEYSKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFL 247
Query: 371 WQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFKVWS 423
W+ + + +PT N+ R I S C C ET H+ C + +W+
Sbjct: 246 WRCISNSLPTAANMRSRHISKDGS---CSRCGMESETVNHILFQCPYARLIWA 97
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.141 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,289,687
Number of Sequences: 36976
Number of extensions: 326679
Number of successful extensions: 2097
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 2062
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2083
length of query: 430
length of database: 9,014,727
effective HSP length: 99
effective length of query: 331
effective length of database: 5,354,103
effective search space: 1772208093
effective search space used: 1772208093
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146564.8


