

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
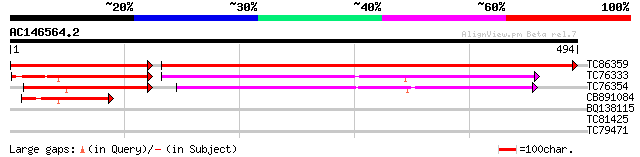
Query= AC146564.2 - phase: 0
(494 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86359 similar to GP|2414681|emb|CAB16318.1 cysteine proteinase... 764 0.0
TC76333 similar to SP|P49044|VPE_VICSA Vacuolar processing enzym... 134 9e-65
TC76354 similar to GP|4589396|dbj|BAA76744.1 asparaginyl endopep... 135 3e-58
CB891084 similar to PIR|T05302|T053 vacuolar processing enzyme (... 80 2e-15
BQ138115 homologue to GP|1431276|emb| ORF YDL173w {Saccharomyces... 30 2.1
TC81425 similar to PIR|T45892|T45892 dTDP-glucose 4-6-dehydratas... 29 3.6
TC79471 similar to GP|21536629|gb|AAM60961.1 seed maturation-lik... 29 4.7
>TC86359 similar to GP|2414681|emb|CAB16318.1 cysteine proteinase precursor
{Vicia narbonensis}, partial (95%)
Length = 1938
Score = 764 bits (1974), Expect(2) = 0.0
Identities = 362/362 (100%), Positives = 362/362 (100%)
Frame = +2
Query: 133 LHWRQRDS*EPIRCYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCI 192
LHWRQRDS*EPIRCYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCI
Sbjct: 521 LHWRQRDS*EPIRCYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCI 700
Query: 193 CYGFYRRFEEETCIWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNIL 252
CYGFYRRFEEETCIWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNIL
Sbjct: 701 CYGFYRRFEEETCIWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNIL 880
Query: 253 SWGGTCPTSRVHHLPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSC 312
SWGGTCPTSRVHHLPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSC
Sbjct: 881 SWGGTCPTSRVHHLPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSC 1060
Query: 313 DAIW*YQHYR*KAIFVPRF*SCNGEPSSTQ*QIRI*NGSCKPKRCRDFIHVGNV*EIRSS 372
DAIW*YQHYR*KAIFVPRF*SCNGEPSSTQ*QIRI*NGSCKPKRCRDFIHVGNV*EIRSS
Sbjct: 1061DAIW*YQHYR*KAIFVPRF*SCNGEPSSTQ*QIRI*NGSCKPKRCRDFIHVGNV*EIRSS 1240
Query: 373 NGKEKRNP*EDCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLK 432
NGKEKRNP*EDCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLK
Sbjct: 1241NGKEKRNP*EDCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLK 1420
Query: 433 IKGSVV*NSLWVTDPVWDETHASICKHLQQWYFRGFYGEGMHGSMWWV*IRAITSIKQSL 492
IKGSVV*NSLWVTDPVWDETHASICKHLQQWYFRGFYGEGMHGSMWWV*IRAITSIKQSL
Sbjct: 1421IKGSVV*NSLWVTDPVWDETHASICKHLQQWYFRGFYGEGMHGSMWWV*IRAITSIKQSL 1600
Query: 493 *C 494
*C
Sbjct: 1601*C 1606
Score = 260 bits (664), Expect(2) = 0.0
Identities = 124/124 (100%), Positives = 124/124 (100%)
Frame = +3
Query: 1 MFSIVLSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGS 60
MFSIVLSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGS
Sbjct: 147 MFSIVLSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGS 326
Query: 61 SGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYV 120
SGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYV
Sbjct: 327 SGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYV 506
Query: 121 GVPK 124
GVPK
Sbjct: 507 GVPK 518
>TC76333 similar to SP|P49044|VPE_VICSA Vacuolar processing enzyme precursor
(EC 3.4.22.-) (VPE) (Proteinase B). [Spring vetch
Tare], partial (91%)
Length = 2253
Score = 134 bits (338), Expect(2) = 9e-65
Identities = 73/130 (56%), Positives = 91/130 (69%), Gaps = 7/130 (5%)
Frame = +3
Query: 2 FSIVLSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVV-------DDAEVDEVGTRWA 54
FS +L +L ++LT+ V+ L D IRLP + + + ++ GTRWA
Sbjct: 9 FSTIL----FLTVILTIFAAVSGSRDLPGD-YIRLPSQSQASRFFHEPENDDNDQGTRWA 173
Query: 55 VLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQ 114
+L+AGS+GY NYRHQADVCHAYQLL KGG+KEENI+VFMYDDIA+N NPRPGVIIN P
Sbjct: 174 ILLAGSNGYWNYRHQADVCHAYQLLRKGGLKEENIIVFMYDDIASNVENPRPGVIINKPD 353
Query: 115 GPNVYVGVPK 124
G +VY GVPK
Sbjct: 354 GGDVYEGVPK 383
Score = 130 bits (328), Expect(2) = 9e-65
Identities = 101/332 (30%), Positives = 161/332 (48%), Gaps = 3/332 (0%)
Frame = +2
Query: 133 LHWRQRDS*EPIRCYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCI 192
LHW + *+ + C++W+ S W+W+ + IL WR SW A++++ +
Sbjct: 386 LHWCRGTC*QFLCCFTWK*ISSYRWEWESCG*WSQ*SYFCILH*SWRSRGSWYARWSLLV 565
Query: 193 CYGFYRRFEEETCIWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNIL 252
C EE+TC W + + + +* W ++* ++ LCDN +C+R+ LGNIL
Sbjct: 566 CI*SE*SLEEKTCFWII*EPSILSGGM*IWQYI*RTSSRRYQYLCDNGFKCSRKQLGNIL 745
Query: 253 SWGGTCPTSRVHHLPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSC 312
WG P+SRV +L + CLDGRQ* Q I +G*R D+ Q + SC
Sbjct: 746 PWGVPSPSSRVLNLLR*PIQYCLDGRQ*HTQFTN*KFAPAI*IG*R*DY---QWLLWFSC 916
Query: 313 DAIW*YQHYR*KAIFVPRF*SCNGEPSSTQ*---QIRI*NGSCKPKRCRDFIHVGNV*EI 369
D +W* + + ++ + + SC + * +I I S PK C +G V +
Sbjct: 917 DGVW*CRA*QQSSLPIFGYKSCQ*QY*FCG*KLLEIEIAFNSS*PKGC*SHSFLG*VPQS 1096
Query: 370 RSSNGKEKRNP*EDCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LG 429
EK + G ++ R C DW +W K + ++ + W T+C**LG
Sbjct: 1097T*GFSAEK*STERSFGSNVPQDACRQQCETDWKALIWH*KGY*TARQC*TCWITTC**LG 1276
Query: 430 MLKIKGSVV*NSLWVTDPVWDETHASICKHLQ 461
+ + G *++LW+ VW ET+ +C+HLQ
Sbjct: 1277LPQNHGENF*DTLWIPISVWYETYEVLCEHLQ 1372
>TC76354 similar to GP|4589396|dbj|BAA76744.1 asparaginyl endopeptidase
(VmPE-1) {Vigna mungo}, partial (91%)
Length = 1660
Score = 135 bits (339), Expect(2) = 3e-58
Identities = 65/115 (56%), Positives = 84/115 (72%), Gaps = 3/115 (2%)
Frame = +2
Query: 13 LLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDE---VGTRWAVLVAGSSGYGNYRHQ 69
L++ TL + + D +RLP + + D+ GT+WA+L+AGS+GY NYRHQ
Sbjct: 44 LIIATLIPIFSAATATAGDDFLRLPSQASRFFQSDDDNNEGTKWAILIAGSNGYWNYRHQ 223
Query: 70 ADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
+DVCHAYQ+L KGG+KEENI+VFMYDDIA+N+ NPRPGVIIN P G +VY GVPK
Sbjct: 224 SDVCHAYQVLRKGGLKEENIIVFMYDDIADNQENPRPGVIINSPHGDDVYKGVPK 388
Score = 108 bits (270), Expect(2) = 3e-58
Identities = 98/318 (30%), Positives = 151/318 (46%), Gaps = 3/318 (0%)
Frame = +1
Query: 146 CYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCICYGFYRRFEEETC 205
C++W+ S W+W+G I+IL R WS+ DA +++ + EEE C
Sbjct: 430 CFTWK*VSSYWWQWKGCG*WS**SHIYIL**SRRSWSAGDAYWSIYVRD*SN*SLEEEAC 609
Query: 206 IWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNILSWGGTCPTSRVHH 265
L K + + W ++*G + S LC+N +C R+ LGNILSWG + + R+ +
Sbjct: 610 F*NL*KPSILSRGMRIWEYL*GSSS*RSEYLCNNSRKCRRKQLGNILSWGES*SSPRI*N 789
Query: 266 LPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSCDAIW*YQHYR*KA 325
+ + CCLDGRQ* Q N I +G R DFK ++ I + DAIW*++ + +
Sbjct: 790 MLG*PIQCCLDGRQ*HTQFANRNFAPAI*IGQRKDFK-RKFNIWFARDAIW*HRAKQG*S 966
Query: 326 IFVPRF*SCNGEPSSTQ*QI---RI*NGSCKPKRCRDFIHVGNV*EIRSSNGKEKRNP*E 382
I + SC + *++ NG P CR +G V + S E +
Sbjct: 967 IPIFGIKSC**KFHFYG*KLISATFKNG--*PA*CRSHPFLG*VPQSSSGFS*ESCSSEA 1140
Query: 383 DCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLKIKGSVV*NSL 442
G + ++R W +W KR + + W T C *LG+ +I G *+ L
Sbjct: 1141SSGSNVSQNAYRREHKTCWKTFIWHEKRSRGACQC*TSWATCCR*LGLPEIIG*DF*DIL 1320
Query: 443 WVTDPVWDETHASICKHL 460
W++ VWDETH +C+ L
Sbjct: 1321WISVSVWDETHEVLCELL 1374
>CB891084 similar to PIR|T05302|T053 vacuolar processing enzyme (EC 3.4.22.-)
isozyme gamma precursor - Arabidopsis thaliana, partial
(11%)
Length = 537
Score = 80.1 bits (196), Expect = 2e-15
Identities = 42/87 (48%), Positives = 57/87 (65%), Gaps = 7/87 (8%)
Frame = +3
Query: 11 WLLLLLTLDGLVARPNHLEWDPVIRLPGEVV-------DDAEVDEVGTRWAVLVAGSSGY 63
W +++T DG + + IRLP + + + ++ GTRWA+L+AGS+GY
Sbjct: 288 WPTMVVTGDGELG----FDSGDYIRLPSQSQASRFFHEPENDDNDQGTRWAILLAGSNGY 455
Query: 64 GNYRHQADVCHAYQLLIKGGVKEENIV 90
NYRHQADVCHAYQLL KGG+KEENI+
Sbjct: 456 WNYRHQADVCHAYQLLRKGGLKEENII 536
>BQ138115 homologue to GP|1431276|emb| ORF YDL173w {Saccharomyces
cerevisiae}, partial (4%)
Length = 338
Score = 30.0 bits (66), Expect = 2.1
Identities = 10/20 (50%), Positives = 12/20 (60%)
Frame = +1
Query: 172 HILLRPWRPWSSWDAKYAVC 191
H L RPWR W W Y++C
Sbjct: 253 HTLYRPWRRWQHWLR*YSLC 312
>TC81425 similar to PIR|T45892|T45892 dTDP-glucose 4-6-dehydratase-like
protein - Arabidopsis thaliana, partial (35%)
Length = 756
Score = 29.3 bits (64), Expect = 3.6
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = +3
Query: 252 LSWGGTCPTSRVHHLPWG 269
L W G+ P +RVH L WG
Sbjct: 240 LGWSGSTPATRVHPLEWG 293
>TC79471 similar to GP|21536629|gb|AAM60961.1 seed maturation-like protein
{Arabidopsis thaliana}, partial (50%)
Length = 1464
Score = 28.9 bits (63), Expect = 4.7
Identities = 15/67 (22%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Frame = +3
Query: 68 HQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGV---IINHPQGPNVYVGVPK 124
H H Y +++ +VV + +A + ++ +P + I N+P P+++ +P
Sbjct: 21 HTQQRFHRYSVVV--------VVVLLLPSMAASTISSQPFILSRITNNPSHPSLHHPLPS 176
Query: 125 IFFFNKP 131
+FF +P
Sbjct: 177 LFFLRRP 197
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.348 0.155 0.614
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,713,729
Number of Sequences: 36976
Number of extensions: 367248
Number of successful extensions: 3008
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 2186
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 764
Number of HSP's gapped (non-prelim): 2329
length of query: 494
length of database: 9,014,727
effective HSP length: 100
effective length of query: 394
effective length of database: 5,317,127
effective search space: 2094948038
effective search space used: 2094948038
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146564.2


