

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.9 + phase: 0
(620 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
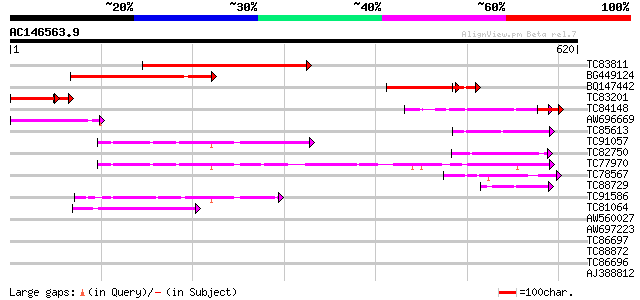
Score E
Sequences producing significant alignments: (bits) Value
TC83811 weakly similar to GP|15983787|gb|AAL10490.1 AT3g18370/MY... 365 e-101
BG449124 weakly similar to GP|15983787|gb AT3g18370/MYF24_8 {Ara... 259 2e-69
BQ147442 similar to GP|11994100|dbj gene_id:MYF24.9~unknown prot... 116 3e-32
TC83201 112 1e-29
TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unk... 78 1e-14
AW696669 61 1e-09
TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta ... 60 2e-09
TC91057 similar to PIR|E86334|E86334 hypothetical protein AAF799... 60 2e-09
TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsi... 60 2e-09
TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {... 59 6e-09
TC78567 similar to PIR|T04143|T04143 CLB1 protein - tomato, part... 51 2e-06
TC88729 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unk... 48 1e-05
TC91586 similar to GP|22655178|gb|AAM98179.1 Ca2+-dependent lipi... 48 1e-05
TC81064 similar to GP|15451210|gb|AAK96876.1 Unknown protein {Ar... 45 1e-04
AW560027 similar to GP|20466318|gb unknown protein {Arabidopsis ... 41 0.001
AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate tr... 40 0.003
TC86697 homologue to GP|3860331|emb|CAA10133.1 hypothetical prot... 39 0.006
TC88872 similar to PIR|T50526|T50526 CaLB protein - Arabidopsis ... 37 0.029
TC86696 similar to GP|3860331|emb|CAA10133.1 hypothetical protei... 36 0.038
AJ388812 weakly similar to GP|15810203|g AT5g50170/K6A12_3 {Arab... 36 0.038
>TC83811 weakly similar to GP|15983787|gb|AAL10490.1 AT3g18370/MYF24_8
{Arabidopsis thaliana}, partial (19%)
Length = 556
Score = 365 bits (938), Expect = e-101
Identities = 184/185 (99%), Positives = 185/185 (99%)
Frame = +2
Query: 146 FSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSL 205
FSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSL
Sbjct: 2 FSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSL 181
Query: 206 HIKGDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDT 265
HIKGDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDT
Sbjct: 182 HIKGDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDT 361
Query: 266 LVKTMVEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSE 325
LVKTMVEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKAS+RQQSGSTNGSSE
Sbjct: 362 LVKTMVEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASKRQQSGSTNGSSE 541
Query: 326 DVSDD 330
DVSDD
Sbjct: 542 DVSDD 556
>BG449124 weakly similar to GP|15983787|gb AT3g18370/MYF24_8 {Arabidopsis
thaliana}, partial (15%)
Length = 536
Score = 259 bits (663), Expect = 2e-69
Identities = 125/160 (78%), Positives = 144/160 (89%)
Frame = +2
Query: 67 YGRYQRKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIV 126
YG YQRK+L EDL+KKW RII+N SP+TPLE CEWLN LL++IW NYFNPKLS+RLSAIV
Sbjct: 50 YGNYQRKILEEDLNKKWNRIIVNTSPVTPLEQCEWLNLLLSQIWSNYFNPKLSTRLSAIV 229
Query: 127 EARLKLRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILL 186
E RLKLRKPRF+ERVE+QEFSLGS PPSL LQG+RWST GDQR++++GFDWDT EMSIL+
Sbjct: 230 EKRLKLRKPRFIERVEVQEFSLGSRPPSLGLQGIRWSTSGDQRLLKMGFDWDTSEMSILM 409
Query: 187 LAKLAKPLMGTARIVINSLHIKGDLIFTPILDGKALLYSF 226
+AKL+ +GTARIVINSLHIKGDL+ TPILDGKALLYSF
Sbjct: 410 VAKLS---VGTARIVINSLHIKGDLLVTPILDGKALLYSF 520
>BQ147442 similar to GP|11994100|dbj gene_id:MYF24.9~unknown protein
{Arabidopsis thaliana}, partial (8%)
Length = 307
Score = 116 bits (290), Expect(2) = 3e-32
Identities = 55/81 (67%), Positives = 66/81 (80%)
Frame = +3
Query: 413 DSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLN 472
DSG+IAK A+FCG+E+EM+VPFEG NS ELKV IVVKEWQFSDG+HSL NL + Q+SL
Sbjct: 3 DSGVIAKHAKFCGEEVEMLVPFEGANSAELKVRIVVKEWQFSDGSHSLTNLHASPQKSLK 182
Query: 473 GSSNIQLRTGKKLKITVVEGK 493
GSSN+ +TG+KLKIT K
Sbjct: 183 GSSNLLSKTGRKLKITCCGSK 245
Score = 40.8 bits (94), Expect(2) = 3e-32
Identities = 21/30 (70%), Positives = 24/30 (80%)
Frame = +1
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYG 514
L+ VVE KDL A K++ GKFDPYIKLQYG
Sbjct: 220 LR*PVVEAKDLDA-KDRFGKFDPYIKLQYG 306
>TC83201
Length = 402
Score = 112 bits (281), Expect(2) = 1e-29
Identities = 54/54 (100%), Positives = 54/54 (100%)
Frame = +2
Query: 1 MSRKKRVFSIDSIEEVAVDFFNYVLQEKPKIPFFIPVILIACAVEKWVFSFSTW 54
MSRKKRVFSIDSIEEVAVDFFNYVLQEKPKIPFFIPVILIACAVEKWVFSFSTW
Sbjct: 182 MSRKKRVFSIDSIEEVAVDFFNYVLQEKPKIPFFIPVILIACAVEKWVFSFSTW 343
Score = 35.4 bits (80), Expect(2) = 1e-29
Identities = 16/21 (76%), Positives = 17/21 (80%)
Frame = +3
Query: 49 FSFSTWVPLALAVWATIQYGR 69
F F VPLALAVWATIQYG+
Sbjct: 327 FLFLLGVPLALAVWATIQYGK 389
>TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unknown
protein {Arabidopsis thaliana}, partial (16%)
Length = 840
Score = 77.8 bits (190), Expect = 1e-14
Identities = 53/164 (32%), Positives = 83/164 (50%), Gaps = 1/164 (0%)
Frame = +1
Query: 432 VPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVE 491
+P EG +SGEL++ I W N + S S + T +++ ++E
Sbjct: 7 IPLEGVSSGELRLKIEAI-WV------------ENQEGSKGPPSGV---TNGWIELVLIE 138
Query: 492 GKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEVGGGEYLKLKVF 550
+DL AA + G DP++++ YG + ++TK H T NP W+QT+EF + G L +K
Sbjct: 139 ARDLIAA-DLRGTSDPFVRVNYGNLKKRTKVVHKTINPRWDQTLEFLDDGSPLTLHVKDH 315
Query: 551 TEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKI 594
L +IG V + L D WIPL+ V+SGEI ++I
Sbjct: 316 NA-LLPTSSIGECVVEYQSLPPNQTSDKWIPLQGVKSGEIHIQI 444
Score = 46.2 bits (108), Expect = 4e-05
Identities = 22/28 (78%), Positives = 25/28 (88%)
Frame = +1
Query: 578 VWIPLERVRSGEIRLKIEAIKVDDQEGS 605
VWIPLE V SGE+RLKIEAI V++QEGS
Sbjct: 1 VWIPLEGVSSGELRLKIEAIWVENQEGS 84
>AW696669
Length = 491
Score = 60.8 bits (146), Expect = 1e-09
Identities = 35/109 (32%), Positives = 60/109 (54%), Gaps = 6/109 (5%)
Frame = +2
Query: 1 MSRKKRVFSIDSIEEVAVDFFNYVLQ--EKPKIPFFIPVILIACAVEKWVFSFSTWVPLA 58
MS KK + +++EE +V N ++ E +I +F+ ++ IA + KW+FSFS +P+
Sbjct: 173 MSSKKGLMIPNNLEEASVKLLNQFVKVKENSRISYFLILVFIAWFIHKWIFSFSNCLPVI 352
Query: 59 LAVWATIQYGRYQRKLLVEDLDKKWKRIILNNSPITPLEH----CEWLN 103
L ++A+ QYG Y + L+++ + +S L H CEWLN
Sbjct: 353 LLLFASTQYGNYHEENT*RGLEQEMESY---HSQYLRLSHHWEQCEWLN 490
>TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta {Homo
sapiens}, partial (3%)
Length = 584
Score = 60.5 bits (145), Expect = 2e-09
Identities = 45/117 (38%), Positives = 65/117 (55%), Gaps = 6/117 (5%)
Frame = +1
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGK-VMQKTKTSH-TPNPVWNQTIEFDEVGGG 542
LK+ VV GK L ++ GK DPY++L K Q+T T T NP WN+T F+ V G
Sbjct: 91 LKVIVVRGKKLKD-EDAIGKSDPYVELWIKKDYKQRTTTKKGTVNPEWNETFTFN-VDGD 264
Query: 543 EYLKLKVFTEELFGDENIGSAQVNLEGLVDGSV--RDVWIP--LERVRSGEIRLKIE 595
+L LKV +++ D+ IG A+V+L+ + D DV +P L G I+L +E
Sbjct: 265 HHLYLKVLDDDVVSDDKIGEAKVDLKQVYDSHYLSTDVKLPALLGLTNHGFIQLVLE 435
>TC91057 similar to PIR|E86334|E86334 hypothetical protein AAF79904.1
[imported] - Arabidopsis thaliana, partial (42%)
Length = 1039
Score = 60.5 bits (145), Expect = 2e-09
Identities = 52/243 (21%), Positives = 107/243 (43%), Gaps = 6/243 (2%)
Frame = +2
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRF-LERVELQEFSLGSCPPSL 155
+ +WLNK + +WP Y N + I + + + P++ ++ VE +E +LGS PP+
Sbjct: 296 DRLDWLNKFVECMWP-YLNKAICKTTRTIAKPIIAEQIPKYKIDSVEFEELNLGSLPPT- 469
Query: 156 ALQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIFT 214
QGM+ +ST + +M+L W + +++A A L T ++V + ++
Sbjct: 470 -FQGMKVYSTDEKELIMELSMKWAGNPN--IIVAVKAFGLRATVQVVDLQVFASPRIMLK 640
Query: 215 PILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTM 270
P++ + S + P V G+ + S+ PG+ ++++ D + K
Sbjct: 641 PLVPSFPCFANIYVSLMEKPHVDFGLKLLGADAMSI-----PGLYRIVQEIIKDQVAKMY 805
Query: 271 VEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVSDD 330
+ P+ + K GI++V+++ A KL + + + N ++
Sbjct: 806 LWPKALQVQIMDPSQAMKKPVGILHVKILKAVKLRKERYNGEEQTLM*NLNSKMINLLQR 985
Query: 331 KDL 333
K L
Sbjct: 986 KQL 994
>TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsis
thaliana}, partial (95%)
Length = 623
Score = 60.1 bits (144), Expect = 2e-09
Identities = 43/113 (38%), Positives = 64/113 (56%), Gaps = 3/113 (2%)
Frame = +3
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTP-NPVWNQTIEFDEVGGG 542
+LK+ VV+GK L KT DPY+ L+ G KTK ++ NPVWN+ + F
Sbjct: 81 QLKVIVVQGKRLVIRDFKTS--DPYVVLKLGNQTAKTKVINSCLNPVWNEELNFTLTEPL 254
Query: 543 EYLKLKVFTEELF-GDENIGSAQVNLEGLVDGS-VRDVWIPLERVRSGEIRLK 593
L L+VF ++L D+ +G+A +NL+ LV + +RD+ RV SGE L+
Sbjct: 255 GVLNLEVFDKDLLKADDKMGNAFINLQPLVSAARLRDIL----RVSSGEQTLR 401
>TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {Arabidopsis
thaliana}, partial (97%)
Length = 2062
Score = 58.9 bits (141), Expect = 6e-09
Identities = 103/523 (19%), Positives = 216/523 (40%), Gaps = 24/523 (4%)
Frame = +2
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRF-LERVELQEFSLGSCPPSL 155
+ +WLNK + +WP Y + + I + ++ + P++ ++ VE Q +LG+ PP+
Sbjct: 377 DRVDWLNKFIELMWP-YLDKAICKTAKNIAKPIIEEQIPKYKIDSVEFQTLTLGTLPPT- 550
Query: 156 ALQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIFT 214
QGM+ + T + +M+ W + + +A A L T ++V + + +
Sbjct: 551 -FQGMKVYVTDEKELIMEPSIKWAGNPN--VTIAVKAFGLKATVQVVDLQVFLLPRITLK 721
Query: 215 PILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTM 270
P++ + + + P V G+ S+ PGV +++L D +
Sbjct: 722 PLVPSFPCFANIYVALMEKPHVDFGLKLLGADLMSI-----PGVYRIVQELIKDQVANMY 886
Query: 271 VEPRRRCFTLPAVDLRK--KAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVS 328
+ P+ + +D+ K + GI++V+V+ A KL +D+
Sbjct: 887 LWPKN--LEVQILDMAKAMRRPVGILHVKVLHAMKL------------------KKKDLL 1006
Query: 329 DDKDLHTFVEVEIEEL-TRRTDVR-LGSTPRWDAPFNMVLHD-NTGTLRFNLYECIPNNV 385
D + +++ +++ +++T V+ P W+ FN+V+ D T L+ N+Y+
Sbjct: 1007GASDPYVKLKLTDDKMPSKKTTVKHKNLNPEWNEEFNLVVKDPETQVLQLNVYDW-EQVG 1183
Query: 386 KCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNS---GEL 442
K D +G I L+ V + + +F D ++ + P + N G++
Sbjct: 1184KHDKMGMNVITLKEVSPE---------------EPKRFTLDLLKTMDPNDAQNEKSRGQI 1318
Query: 443 KVSIVVK---EWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAK 499
V + K E + G + + + G G +L + V E +D+
Sbjct: 1319VVEVTYKPLNEEEMGKGFDETQTIPKAPEGTPAG--------GGQLVVIVHEAQDV---- 1462
Query: 500 EKTGKFDPYIKLQYGKVMQKTK-TSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEE--- 553
E +P +L + +KTK +P W +F +E + L ++V +
Sbjct: 1463EGKHHTNPQARLIFRGEEKKTKRIKKNRDPRWEDEFQFIAEEPPTNDKLHVEVVSSSSRT 1642
Query: 554 -LFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
L E++G +NL +V + L ++G I+++++
Sbjct: 1643LLHQKESLGYVDINLGDVVSNKRINEKYHLIDSKNGRIQVELQ 1771
>TC78567 similar to PIR|T04143|T04143 CLB1 protein - tomato, partial (53%)
Length = 1159
Score = 50.8 bits (120), Expect = 2e-06
Identities = 41/135 (30%), Positives = 73/135 (53%), Gaps = 6/135 (4%)
Frame = +3
Query: 475 SNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT---SHTPNPVWN 531
S+++L+ LK+T+V+ DL E GK DPY+ L Y + + K KT ++ NPVW+
Sbjct: 150 SDLELKPHGSLKVTIVKATDLKNM-EMIGKSDPYVVL-YIRPLFKVKTKVINNNLNPVWD 323
Query: 532 QTIE-FDEVGGGEYLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEI 590
QT E E + L L+VF E++ D+ +G ++ L + + ++ + E+
Sbjct: 324 QTFELIAEDKETQSLILEVFDEDIGQDKRLGIVKLPL----------IELEVQTEKELEL 473
Query: 591 RL--KIEAIKVDDQE 603
RL ++ +KV D++
Sbjct: 474 RLLSSLDTLKVKDKK 518
>TC88729 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1047
Score = 48.1 bits (113), Expect = 1e-05
Identities = 30/80 (37%), Positives = 41/80 (50%)
Frame = +1
Query: 515 KVMQKTKTSHTPNPVWNQTIEFDEVGGGEYLKLKVFTEELFGDENIGSAQVNLEGLVDGS 574
KV+ KT T P WNQT+EF + G L +K L +IG V + L
Sbjct: 7 KVIYKTLT-----PQWNQTLEFPDDGSPLMLYVKDHNA-LLPTSSIGECVVEYQRLPPNQ 168
Query: 575 VRDVWIPLERVRSGEIRLKI 594
+ D WIPL+ V+ GEI +K+
Sbjct: 169 MADKWIPLQGVKRGEIHIKL 228
>TC91586 similar to GP|22655178|gb|AAM98179.1 Ca2+-dependent lipid-binding
protein putative {Arabidopsis thaliana}, partial (40%)
Length = 676
Score = 47.8 bits (112), Expect = 1e-05
Identities = 62/238 (26%), Positives = 101/238 (42%), Gaps = 10/238 (4%)
Frame = +1
Query: 72 RKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLK 131
RKLL W + N +T WLN LT+IWP Y N S + E L+
Sbjct: 10 RKLLPSQFYPSWV-VFSNRQKLT------WLNSHLTKIWP-YVNEAASELIKTSAEPILE 165
Query: 132 LRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLA 191
+P L ++ +F+LG+ P + GD M+L WD + IL +
Sbjct: 166 EYRPMILSALKFSKFTLGTVAPQFTGVSI-IEDGGDGVTMELEVQWDGNPSIILDI---- 330
Query: 192 KPLMGTA-RIVINSLHIKG--DLIFTPILDG----KALLYSFVSAPEVRVGVAFGSGGSQ 244
K L+G A + + ++ G LIF P+++ A+ YS ++ + G
Sbjct: 331 KTLVGLALPVQVKNVGFTGVFRLIFKPLVNEFPGFGAVCYSLRQKKKLDFTLKVIGGDIS 510
Query: 245 SLPATEWPGVSSWLEKLFTDTLVKTMVEPRRRCF-TLPA--VDLRKKAVGGIIYVRVI 299
++ PG+ +E D + ++ P R+ LP DL K V GI+ V+++
Sbjct: 511 TI-----PGLYDAIEGAIRDAVEDSITWPVRKIVPILPGDYSDLELKPV-GILXVKLV 666
>TC81064 similar to GP|15451210|gb|AAK96876.1 Unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 928
Score = 44.7 bits (104), Expect = 1e-04
Identities = 41/144 (28%), Positives = 69/144 (47%), Gaps = 4/144 (2%)
Frame = +2
Query: 69 RYQRKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSR-LSAIVE 127
R ++KL E+ + +R +L++S E WLN + IWP S + L I+
Sbjct: 506 RLRKKLQFEERKQSNQRRVLSDS-----ETVRWLNHAVENIWPICMEQIASQKILLPIIP 670
Query: 128 ARLKLRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDT-HEMSILL 186
L+ KP + +Q LG PP + + D V++LG ++ T +MS +L
Sbjct: 671 WFLEKYKPWTAKEASVQHLYLGRNPPLITDIRVLRQCDDDHLVLELGMNFLTGDDMSAIL 850
Query: 187 LAKLAKPL-MG-TARIVINSLHIK 208
KL K L G +A++ I +H++
Sbjct: 851 AVKLRKRLGFGMSAKLHITGMHVE 922
>AW560027 similar to GP|20466318|gb unknown protein {Arabidopsis thaliana},
partial (24%)
Length = 585
Score = 41.2 bits (95), Expect = 0.001
Identities = 38/143 (26%), Positives = 66/143 (45%), Gaps = 6/143 (4%)
Frame = +2
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLA 156
E EW+N +L ++W Y + + + L+KP ++ERVE+++FSLG P S+
Sbjct: 23 ESVEWVNMVLGKLWKVYRGGLENWIIGLLQPVIDDLKKPDYVERVEIKQFSLGDEPLSVR 202
Query: 157 LQGMRWSTIGDQRVMQLGFDWDTHEMSILLLA------KLAKPLMGTARIVINSLHIKGD 210
R S + Q+G + +L L+ + P+ + L +K
Sbjct: 203 NVERRTSRRVNDLQYQIGLRYTGGARMLLSLSLKFGIIPIVVPVGVRDFDIDGELWVKLR 382
Query: 211 LIFTPILDGKALLYSFVSAPEVR 233
LI T G A ++FVS P+++
Sbjct: 383 LIPTAPWVG-AASWAFVSLPKIK 448
>AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(5%)
Length = 778
Score = 40.0 bits (92), Expect = 0.003
Identities = 27/88 (30%), Positives = 49/88 (55%), Gaps = 1/88 (1%)
Frame = +3
Query: 478 QLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTS-HTPNPVWNQTIEF 536
++ T +KL + V++ ++LA K+ G PYI + + +KT+T NPVWN+T+ F
Sbjct: 135 KMATVRKLIVEVIDAQNLAP-KDGHGTSSPYIVIDFHGQRRKTRTLVRDLNPVWNETLSF 311
Query: 537 DEVGGGEYLKLKVFTEELFGDENIGSAQ 564
+ VG + V+ +++ D G A+
Sbjct: 312 N-VGERNEIFGDVWELDVYHDMKHGPAR 392
>TC86697 homologue to GP|3860331|emb|CAA10133.1 hypothetical protein {Cicer
arietinum}, partial (89%)
Length = 825
Score = 38.9 bits (89), Expect = 0.006
Identities = 28/106 (26%), Positives = 47/106 (43%), Gaps = 8/106 (7%)
Frame = +2
Query: 475 SNIQLRTGKK-----LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTP--N 527
S +Q + +K L++ ++ K L + DPY+ L Y K+ N
Sbjct: 107 SKLQFQNQRKMPRGTLEVILISAKGLDD-NDFLSSIDPYVILSYSGQEHKSTVQEGAGSN 283
Query: 528 PVWNQTIEFDEVGGGEYLKLKVFTEELF-GDENIGSAQVNLEGLVD 572
P WN+T F L LK+ ++ + D+NIG A + LE + +
Sbjct: 284 PQWNETFLFTVSDNASELNLKIMEKDNYNNDDNIGEAIIPLEAVFE 421
>TC88872 similar to PIR|T50526|T50526 CaLB protein - Arabidopsis thaliana,
partial (35%)
Length = 699
Score = 36.6 bits (83), Expect = 0.029
Identities = 26/95 (27%), Positives = 48/95 (50%)
Frame = +3
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLA 156
E +WLNKLL+++WP + + + VE L+ +P + ++ + SLG+ P +
Sbjct: 345 EQVKWLNKLLSKLWP-FVAEAATMVIKESVEPLLEEYRPPGITSLKFSKLSLGNVAPKIE 521
Query: 157 LQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLA 191
++ T G Q +M + W + SI+L + A
Sbjct: 522 SIRVQSLTKG-QIIMDVDLRWG-GDPSIILAVEAA 620
>TC86696 similar to GP|3860331|emb|CAA10133.1 hypothetical protein {Cicer
arietinum}, partial (90%)
Length = 733
Score = 36.2 bits (82), Expect = 0.038
Identities = 26/95 (27%), Positives = 44/95 (45%), Gaps = 4/95 (4%)
Frame = +1
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTP--NPVWNQTIEFDEVGGG 542
L++ ++ K L + DPY+ L Y K+ NP WN+T F
Sbjct: 100 LEVVLISAKGLED-NDFLSSIDPYVILTYRAQEHKSTVQEGAGSNPQWNETFLFTVSDTA 276
Query: 543 EYLKLKVFTEELF-GDENIGSAQVNLEGLV-DGSV 575
L LK+ ++ + D+N+G + LE ++ +GSV
Sbjct: 277 YELNLKIMEKDNYSADDNLGEVIIPLETVIQEGSV 381
>AJ388812 weakly similar to GP|15810203|g AT5g50170/K6A12_3 {Arabidopsis
thaliana}, partial (14%)
Length = 673
Score = 36.2 bits (82), Expect = 0.038
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Frame = +2
Query: 482 GKKLKITVVEGKDLAAAKEKTGKFDPYIKLQY-GKVMQKTKTSHTPNPVWNQTIEFD 537
G + + ++EG DL + E TG DPY+ G+ + T +P WN+ +EFD
Sbjct: 278 GWVVTVALIEGVDLVSL-ESTGLSDPYVVFTCNGQTRSSSVKLETSDPQWNEILEFD 445
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,523,602
Number of Sequences: 36976
Number of extensions: 279385
Number of successful extensions: 1399
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 1380
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1390
length of query: 620
length of database: 9,014,727
effective HSP length: 102
effective length of query: 518
effective length of database: 5,243,175
effective search space: 2715964650
effective search space used: 2715964650
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146563.9


