

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.12 - phase: 0
(336 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
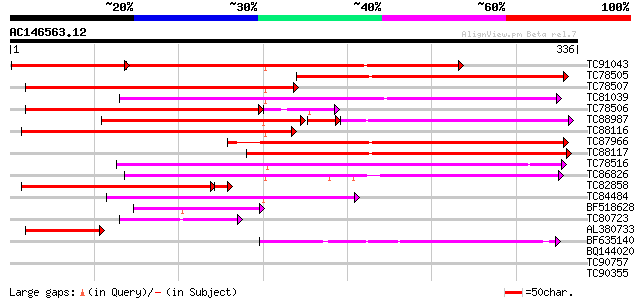
Score E
Sequences producing significant alignments: (bits) Value
TC91043 similar to GP|15293171|gb|AAK93696.1 putative lipase {Ar... 246 3e-84
TC78505 similar to GP|15293171|gb|AAK93696.1 putative lipase {Ar... 219 9e-58
TC78507 similar to GP|15293171|gb|AAK93696.1 putative lipase {Ar... 216 8e-57
TC81039 similar to PIR|H86244|H86244 lysophospholipase homolog ... 209 1e-54
TC78506 similar to GP|15293171|gb|AAK93696.1 putative lipase {Ar... 199 5e-54
TC88987 weakly similar to PIR|T00552|T00552 lysophospholipase ho... 120 3e-53
TC88116 weakly similar to PIR|E96803|E96803 probable lipase 416... 204 3e-53
TC87966 weakly similar to PIR|E96803|E96803 probable lipase 416... 193 7e-50
TC88117 weakly similar to PIR|E96803|E96803 probable lipase 416... 177 7e-45
TC78516 similar to GP|21554372|gb|AAM63479.1 phospholipase-like ... 163 8e-41
TC86826 similar to PIR|F96568|F96568 probable lipase 20450-2164... 162 1e-40
TC82858 weakly similar to PIR|E96803|E96803 probable lipase 416... 140 1e-35
TC84484 similar to GP|20161418|dbj|BAB90342. phospholipase-like ... 109 2e-24
BF518628 weakly similar to PIR|T00552|T005 lysophospholipase hom... 67 1e-11
TC80723 similar to PIR|T48524|T48524 lysophospholipase-like prot... 61 7e-10
AL380733 59 3e-09
BF635140 similar to PIR|G71415|G71 hypothetical protein - Arabid... 45 3e-05
BQ144020 homologue to PIR|B84620|B846 hypothetical protein At2g2... 36 0.018
TC90757 similar to GP|17528996|gb|AAL38708.1 unknown protein {Ar... 36 0.018
TC90355 similar to GP|9294510|dbj|BAB02772.1 gene_id:MDB19.2~unk... 35 0.031
>TC91043 similar to GP|15293171|gb|AAK93696.1 putative lipase {Arabidopsis
thaliana}, partial (67%)
Length = 1121
Score = 246 bits (629), Expect(2) = 3e-84
Identities = 121/213 (56%), Positives = 155/213 (71%), Gaps = 13/213 (6%)
Frame = +2
Query: 70 VFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLH 129
+F K K ++CHGY DTCTF+FEG+ARKLAS G+GVFA+DYPGFGLS+GLH
Sbjct: 485 LFGKVGFLRNLSQKQQYFFCHGYGDTCTFFFEGIARKLASCGYGVFAMDYPGFGLSEGLH 664
Query: 130 GYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWDGAALI 176
GYI SF+ LV+DVIE +SKIK GESMGGA+AL HFKQP AW+GA L
Sbjct: 665 GYITSFDQLVDDVIEQYSKIKENPEFKSLPCFLFGESMGGAVALKAHFKQPKAWNGAILC 844
Query: 177 APLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFY 236
AP+CK +E M+P LV ++LI ++ VLPK KLVP ++ + ++D +KRE YNV+ Y
Sbjct: 845 APMCKISEKMVPPKLVVKMLIAMSNVLPKNKLVP-TNDIGDAAFKDPKKREQTHYNVISY 1021
Query: 237 KDKPRLGTALELLKATQELEQRLEEVSLPLLVM 269
KDKPRL TA+ELLK T+E+EQ+LEEV+LPL ++
Sbjct: 1022KDKPRLRTAVELLKTTEEIEQKLEEVTLPLFIL 1120
Score = 83.2 bits (204), Expect(2) = 3e-84
Identities = 40/70 (57%), Positives = 50/70 (71%)
Frame = +3
Query: 2 LIEMGEIEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIY 61
L M + G+ ELQ + MDE PARR AREAFKDIQL IDH LF+ P DG+ M+E Y
Sbjct: 279 LSRMKRLPGVDNELQKKLDAKMDEVPARRQAREAFKDIQLGIDHILFKTPCDGLXMKESY 458
Query: 62 EVNSRGLKVF 71
EVNS+G+++F
Sbjct: 459 EVNSKGIEIF 488
>TC78505 similar to GP|15293171|gb|AAK93696.1 putative lipase {Arabidopsis
thaliana}, partial (43%)
Length = 630
Score = 219 bits (559), Expect = 9e-58
Identities = 107/161 (66%), Positives = 127/161 (78%)
Frame = +1
Query: 171 DGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAP 230
D A AP+CK A+DM P WL+ QILIG+A VLPK KLVPQK + E +RD +KRE+
Sbjct: 28 DDAIFDAPMCKIADDMAPPWLLAQILIGIANVLPKQKLVPQKN-LAEAAFRDLKKREMTA 204
Query: 231 YNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAK 290
YNV+ YKDKPRL TA+E+LK TQE+E+RLEEVSLPLL++HGEADI+TDPS SK Y+KA
Sbjct: 205 YNVVAYKDKPRLWTAVEMLKTTQEIEKRLEEVSLPLLILHGEADIVTDPSVSKTFYEKAS 384
Query: 291 VKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSS 331
DKKL LYKDA+H+LLEGEPDE I V DII WLD+HSS
Sbjct: 385 SSDKKLKLYKDAYHSLLEGEPDEMIIQVFSDIILWLDEHSS 507
>TC78507 similar to GP|15293171|gb|AAK93696.1 putative lipase {Arabidopsis
thaliana}, partial (43%)
Length = 659
Score = 216 bits (551), Expect = 8e-57
Identities = 104/175 (59%), Positives = 129/175 (73%), Gaps = 13/175 (7%)
Frame = +2
Query: 10 GMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRGLK 69
G+ ELQ I +NMD ARR AREAFKD+QL IDH LF+ DG+KM+E YEVNS+G++
Sbjct: 128 GVDNELQKILDANMDHVSARRQAREAFKDVQLGIDHILFKTQCDGLKMKESYEVNSKGIE 307
Query: 70 VFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLH 129
+F KSW PE + K V+YCHGY DT TF+FEG+ARKLA G+GVFA+DYPGFGLS+GLH
Sbjct: 308 IFYKSWFPETARPKAAVFYCHGYGDTSTFFFEGIARKLACDGYGVFAMDYPGFGLSEGLH 487
Query: 130 GYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWD 171
YIPSF++L++DVIE +SKIK G+SMGGA+AL +H KQP D
Sbjct: 488 CYIPSFDSLIDDVIEIYSKIKENPELQSLPSFLFGQSMGGAVALKMHLKQPKIAD 652
>TC81039 similar to PIR|H86244|H86244 lysophospholipase homolog 25331-24357
[imported] - Arabidopsis thaliana, partial (86%)
Length = 1070
Score = 209 bits (532), Expect = 1e-54
Identities = 106/276 (38%), Positives = 166/276 (59%), Gaps = 14/276 (5%)
Frame = +1
Query: 66 RGLKVFSKSWIPEKS-PMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGL 124
R L +F++SW+P + P + +++ HGY + ++ F+ LA GF F+LD G G
Sbjct: 148 RNLTLFTRSWLPNPTTPPRALIFMIHGYGNDISWTFQSTPIFLAQMGFACFSLDLQGHGH 327
Query: 125 SDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWD 171
S GL ++PS + +V D + F+ +K GESMGGAI+L IHF P +
Sbjct: 328 SQGLKAFVPSVDLVVQDCLSFFNSVKKDSNFFGLPCFLYGESMGGAISLLIHFADPKGFQ 507
Query: 172 GAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPY 231
GA L+AP+CK ++ + P W + QIL +AK P +VP + + +++ D K+ +A
Sbjct: 508 GAILVAPMCKISDKVRPKWPIPQILTFLAKFFPTLPIVPTPDLLYKSVKVD-HKKVIAQM 684
Query: 232 NVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKV 291
N L Y+ KPRLGT +ELL+ T L ++L +V LP +V+HG AD++TDP S+ LY++A+
Sbjct: 685 NPLRYRGKPRLGTVVELLRVTDILSRKLCDVELPFIVLHGSADVVTDPEVSRELYEEARS 864
Query: 292 KDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
DK + ++ H+LL GE DE + V +DI+ WL+
Sbjct: 865 DDKTIKVFDGMMHSLLFGETDENVEIVRNDILQWLN 972
>TC78506 similar to GP|15293171|gb|AAK93696.1 putative lipase {Arabidopsis
thaliana}, partial (45%)
Length = 766
Score = 199 bits (505), Expect(2) = 5e-54
Identities = 92/141 (65%), Positives = 113/141 (79%)
Frame = +3
Query: 10 GMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRGLK 69
G+ ELQ I +NMD ARR AREAFKD+QL IDH LF+ DG+KM+E YEVNS+G++
Sbjct: 150 GVDNELQKILDANMDHVSARRQAREAFKDVQLGIDHILFKTQCDGLKMKESYEVNSKGIE 329
Query: 70 VFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLH 129
+F KSW PE + K V+YCHGY DT TF+FEG+ARKLA G+GVFA+DYPGFGLS+GLH
Sbjct: 330 IFYKSWFPETARPKAAVFYCHGYGDTSTFFFEGIARKLACDGYGVFAMDYPGFGLSEGLH 509
Query: 130 GYIPSFENLVNDVIEHFSKIK 150
YIPSF++LV+DVIE +SKIK
Sbjct: 510 CYIPSFDSLVDDVIEIYSKIK 572
Score = 30.0 bits (66), Expect(2) = 5e-54
Identities = 18/49 (36%), Positives = 25/49 (50%), Gaps = 4/49 (8%)
Frame = +1
Query: 151 GESMGGAIALNIHFKQPTAWDGAALI----APLCKFAEDMIPHWLVKQI 195
G+SMGG+ P G ++ AP+CK A+DM WL+ QI
Sbjct: 613 GQSMGGSCCFK---DAP*TT*GMGMVPIFVAPMCKIADDMGSPWLLAQI 750
>TC88987 weakly similar to PIR|T00552|T00552 lysophospholipase homolog
F12L6.8 - Arabidopsis thaliana, partial (87%)
Length = 1283
Score = 120 bits (301), Expect(3) = 3e-53
Identities = 56/134 (41%), Positives = 82/134 (60%), Gaps = 13/134 (9%)
Frame = +1
Query: 55 VKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGV 114
+K EE + +NSRG+K+ + SWIP K +++ CHGYA C+ + AR+LA+ G+ V
Sbjct: 220 IKYEEGFVLNSRGMKLLATSWIPANENPKALIFMCHGYAMECSITMDSTARRLANGGYAV 399
Query: 115 FALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIALN 161
+ +DY G G S GL G + +F+ +++D + HFS I GESMGGA+AL
Sbjct: 400 YGIDYEGHGKSYGLPGLVKNFDTIIDDCLRHFSSICEKPENKKKMRYLLGESMGGAVALL 579
Query: 162 IHFKQPTAWDGAAL 175
+H K+P WDGA L
Sbjct: 580 LHRKKPQYWDGAIL 621
Score = 102 bits (253), Expect(3) = 3e-53
Identities = 53/138 (38%), Positives = 82/138 (59%)
Frame = +3
Query: 197 IGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELE 256
+ ++KV P ++VP +++ + ++ R+ N +K KPRL T EL + ++E
Sbjct: 687 VHLSKVAPTWQIVPT-QDIIDVAFKVPEVRQQIRANQYCFKGKPRLRTGYELSRVATKIE 863
Query: 257 QRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIF 316
+ L+EVSLP L++HGE D +TDPS SK L++ A KDK L LY +H LL GEP E +
Sbjct: 864 ETLDEVSLPFLILHGEEDRVTDPSVSKQLHEVASSKDKTLKLYPGMWHGLLYGEPPENLE 1043
Query: 317 HVLDDIISWLDDHSSTKN 334
V DI +W+++ N
Sbjct: 1044IVFKDIFNWIEERCQYGN 1097
Score = 25.0 bits (53), Expect(3) = 3e-53
Identities = 10/20 (50%), Positives = 15/20 (75%)
Frame = +2
Query: 177 APLCKFAEDMIPHWLVKQIL 196
AP+CK A+D+ P+ V +IL
Sbjct: 626 APMCKIADDIKPNAFVLRIL 685
>TC88116 weakly similar to PIR|E96803|E96803 probable lipase 4162-5963
[imported] - Arabidopsis thaliana, partial (45%)
Length = 917
Score = 204 bits (520), Expect = 3e-53
Identities = 96/177 (54%), Positives = 131/177 (73%), Gaps = 14/177 (7%)
Frame = +3
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
I+G+SEEL +I N+D A +RR R AF ++Q +DHCLF+ G++ EE YE NSRG
Sbjct: 387 IDGVSEELNDIASYNLDFAYSRRRVRSAFAEVQQQLDHCLFKDAPAGIRAEEWYERNSRG 566
Query: 68 LKVFSKSWIPEKS-PMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
L++F KSW+PE P+K V +CHGY DTCTF+FEGVAR++A+SG+ VFA+DYPGFGLS+
Sbjct: 567 LEIFCKSWMPESGIPIKASVCFCHGYGDTCTFFFEGVARRIAASGYAVFAMDYPGFGLSE 746
Query: 127 GLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAW 170
GLHGYIP+F++LV+DVIEH+++IK G+SMGGA +L ++ K+P W
Sbjct: 747 GLHGYIPNFDDLVDDVIEHYTQIKARPDLRELPRVLLGQSMGGAXSLKVYLKEPNNW 917
>TC87966 weakly similar to PIR|E96803|E96803 probable lipase 4162-5963
[imported] - Arabidopsis thaliana, partial (48%)
Length = 948
Score = 193 bits (491), Expect = 7e-50
Identities = 98/202 (48%), Positives = 134/202 (65%)
Frame = +2
Query: 130 GYIPSFENLVNDVIEHFSKIKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPH 189
GY+P F I G+SMGGAIAL H +P WDG L+AP+CK +E M+P
Sbjct: 8 GYLPQF-------------IFGQSMGGAIALKAHLNEPNVWDGVILVAPMCKISEGMLPP 148
Query: 190 WLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELL 249
+ + L ++K++PK KL P K+ + E I+R+ KR+LA YNV+ Y D+ RL T +ELL
Sbjct: 149 TTILKALTLLSKMMPKAKLFPYKD-LSELIFREPGKRKLAVYNVISYDDQTRLRTGMELL 325
Query: 250 KATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEG 309
ATQ++E +LE+VS PLL++HG D +TDP S+ LY+KA KDK L +Y+ +H +LEG
Sbjct: 326 SATQDIESQLEKVSAPLLILHGAEDKVTDPLVSQFLYEKASSKDKTLKIYEGGYHGILEG 505
Query: 310 EPDETIFHVLDDIISWLDDHSS 331
EPDE I V +DIISWLD+ S
Sbjct: 506 EPDERISSVHNDIISWLDNRCS 571
>TC88117 weakly similar to PIR|E96803|E96803 probable lipase 4162-5963
[imported] - Arabidopsis thaliana, partial (35%)
Length = 775
Score = 177 bits (448), Expect = 7e-45
Identities = 89/193 (46%), Positives = 129/193 (66%)
Frame = +3
Query: 141 DVIEHFSKIKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVA 200
D+ ++ + G+SMGG L + L+AP+CK A+D++P V ++L ++
Sbjct: 12 DLRDYPRVLLGQSMGGRFLLKFI*RSQIIGMLVMLVAPMCKIADDVLPPDAVMKVLTLLS 191
Query: 201 KVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLE 260
KV+PK KL P K+ + E +R+ KR+LAPYNV+ Y+D PRL T +ELL+ T+E+E ++E
Sbjct: 192 KVMPKAKLFPNKD-LAELAFREPSKRKLAPYNVICYEDNPRLKTGMELLRVTKEIESKVE 368
Query: 261 EVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLD 320
+VS PLL++HG AD +TDP SK LY+ A KDK L LY++ +H +LEGEPD+ I V D
Sbjct: 369 KVSAPLLILHGAADKVTDPLVSKFLYENASSKDKTLKLYENGYHCILEGEPDDRIKAVHD 548
Query: 321 DIISWLDDHSSTK 333
DI+SWLD S K
Sbjct: 549 DIVSWLDSRCSVK 587
>TC78516 similar to GP|21554372|gb|AAM63479.1 phospholipase-like protein
{Arabidopsis thaliana}, partial (90%)
Length = 1190
Score = 163 bits (413), Expect = 8e-41
Identities = 97/281 (34%), Positives = 151/281 (53%), Gaps = 14/281 (4%)
Frame = +1
Query: 64 NSRGLKVFSKSWIPEK-SPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGF 122
NSRGLK+F++ WIP + + G + HGY ++ + A A +GF A+D+ G
Sbjct: 94 NSRGLKLFTQWWIPNPPTKLIGTLAVVHGYTGESSWTVQLSAVYFAKAGFATCAIDHQGH 273
Query: 123 GLSDGLHGYIPSFENLVNDVIEHFSKIKG------------ESMGGAIALNIHFKQP-TA 169
G SDGL +IP +V+D I F + ES+GGAIAL I ++
Sbjct: 274 GFSDGLIAHIPDVNPVVDDCISFFESFRSRFDSSLPSFLYSESLGGAIALLITLRRGGLP 453
Query: 170 WDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELA 229
W+G L +C ++ P W ++ L A V+P ++VP + + + +++ KR+LA
Sbjct: 454 WNGLILNGAMCGVSDKFKPPWPLEHFLSLAAAVIPTWRVVPTRGSIPDVSFKEEWKRKLA 633
Query: 230 PYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKA 289
+ +PR TA ELL+ +EL+ R EEV +P L +HG DI+ DP+ + LY +A
Sbjct: 634 IASPKRTVARPRAATAQELLRICRELQGRFEEVDVPFLAVHGGDDIVCDPACVEELYSRA 813
Query: 290 KVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHS 330
KDK L +Y +H L+ GEP+E + V D++ WL H+
Sbjct: 814 GSKDKTLKIYDGMWHQLV-GEPEENVELVFGDMLEWLIKHA 933
>TC86826 similar to PIR|F96568|F96568 probable lipase 20450-21648
[imported] - Arabidopsis thaliana, partial (93%)
Length = 1214
Score = 162 bits (411), Expect = 1e-40
Identities = 98/280 (35%), Positives = 146/280 (52%), Gaps = 20/280 (7%)
Frame = +2
Query: 69 KVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGL 128
K+F++S++P + +K VY HGY + F+ + A+ G+ VF D G G SDGL
Sbjct: 155 KIFTQSFLPLNAEIKATVYMTHGYGSDTGWLFQKICITYATWGYAVFTADLLGHGRSDGL 334
Query: 129 HGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFK-QPTAWDGAA 174
Y+ + + + F ++ GESMGG L ++F+ +P W G
Sbjct: 335 RCYLGDMDKIAATSLSFFLHVRRSPPYNHLPAFLFGESMGGLATLLMYFQSEPDTWTGLI 514
Query: 175 LIAPLCKFAEDMIP---HWLVKQILIGVAKV---LPKTKLVPQKEEVKENIYRDARKREL 228
APL EDM P H V +L G+A +P K+V + RD K ++
Sbjct: 515 FSAPLFVIPEDMKPSKIHLFVYGLLFGLADTWAAMPDNKMVGKA-------IRDPNKLKI 673
Query: 229 APYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQK 288
N Y PR+GT ELL+ TQ ++ V++P L HG AD +T PS+SK LY+K
Sbjct: 674 IASNPRRYTGPPRVGTMRELLRVTQYVQDNFCNVTVPFLTAHGTADGVTCPSSSKLLYEK 853
Query: 289 AKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDD 328
A+ KDK L LY+ +H+L++GEPDE+ VL D+ W+D+
Sbjct: 854 AESKDKTLKLYEGMYHSLIQGEPDESANLVLRDMREWIDE 973
>TC82858 weakly similar to PIR|E96803|E96803 probable lipase 4162-5963
[imported] - Arabidopsis thaliana, partial (31%)
Length = 561
Score = 140 bits (354), Expect(2) = 1e-35
Identities = 62/116 (53%), Positives = 88/116 (75%), Gaps = 1/116 (0%)
Frame = +1
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
IEG+S+EL +I N+D AP+RR R+AF + +DH LF+ G+ +E YE NSRG
Sbjct: 181 IEGISDELNSIASLNLDHAPSRRHVRQAFTHLHQQLDHILFKTAPAGIITQEWYERNSRG 360
Query: 68 LKVFSKSWIPEKS-PMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGF 122
L++F KSW+PE P+KG +++CHGY TCTF+FEG+AR++A+SGFGV+A+D+PGF
Sbjct: 361 LEIFCKSWMPEHGVPIKGALFFCHGYGSTCTFFFEGIARRIAASGFGVYAMDFPGF 528
Score = 26.6 bits (57), Expect(2) = 1e-35
Identities = 10/11 (90%), Positives = 11/11 (99%)
Frame = +2
Query: 122 FGLSDGLHGYI 132
FGLS+GLHGYI
Sbjct: 527 FGLSEGLHGYI 559
>TC84484 similar to GP|20161418|dbj|BAB90342. phospholipase-like protein
{Oryza sativa (japonica cultivar-group)}, partial (38%)
Length = 656
Score = 109 bits (272), Expect = 2e-24
Identities = 59/164 (35%), Positives = 87/164 (52%), Gaps = 14/164 (8%)
Frame = +2
Query: 58 EEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFAL 117
+E Y NSRG+++F+ W+P S K +V+ CHGY C+ + + + KLAS+G+ VF +
Sbjct: 155 QEGYWKNSRGMRLFTCKWLPISSSPKALVFLCHGYGMECSGFMKEIGEKLASAGYAVFGM 334
Query: 118 DYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIALNIHF 164
DY G G S G+ YI F+N+VND + I GESMGGA+A+ +H
Sbjct: 335 DYEGHGHSAGVRCYITKFDNVVNDCSNFYKSICELQEYRGKAKFLYGESMGGAVAVLLHK 514
Query: 165 KQPTAWDGAALI-APLCKFAEDMIPHWLVKQILIGVAKVLPKTK 207
K P AP+CK +E + PH ++G+ K K
Sbjct: 515 KDPFILGWCCFSWAPMCKISEKVKPHSCCC*HVVGIGTYFXKWK 646
>BF518628 weakly similar to PIR|T00552|T005 lysophospholipase homolog F12L6.8
- Arabidopsis thaliana, partial (23%)
Length = 468
Score = 67.0 bits (162), Expect = 1e-11
Identities = 33/106 (31%), Positives = 50/106 (47%), Gaps = 28/106 (26%)
Frame = +3
Query: 74 SWIPEKSPMKGIVYYCHGYADTCTFYFE----------------------------GVAR 105
SWIP K +++ CHGYA C+ + G AR
Sbjct: 9 SWIPANENPKALIFMCHGYAMECSITMDSK*LNFSFFLLLYDIIIILYLN*MNEYIGTAR 188
Query: 106 KLASSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIKG 151
+LA+ G+ V+ +DY G G S GL G + +F+ +++D + HFS I G
Sbjct: 189 RLANGGYAVYGIDYEGHGKSYGLPGLVKNFDTIIDDCLRHFSSICG 326
>TC80723 similar to PIR|T48524|T48524 lysophospholipase-like protein -
Arabidopsis thaliana, partial (27%)
Length = 669
Score = 60.8 bits (146), Expect = 7e-10
Identities = 30/73 (41%), Positives = 43/73 (58%)
Frame = +1
Query: 66 RGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLS 125
R +F +SW P +KGI+ HG + Y + AR+L FGV+A+D+ G G S
Sbjct: 421 RNNALFCRSWFPVYGDLKGIMIIIHGLNEHSGRYAD-FARQLTLCNFGVYAMDWIGHGGS 597
Query: 126 DGLHGYIPSFENL 138
DGLHGY+PS + +
Sbjct: 598 DGLHGYVPSLDQV 636
>AL380733
Length = 312
Score = 58.5 bits (140), Expect = 3e-09
Identities = 28/47 (59%), Positives = 33/47 (69%)
Frame = -2
Query: 10 GMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVK 56
G+ ELQ I +NMD ARR AREAFKD+QL IDH LF+ DG+K
Sbjct: 143 GVDNELQKILDANMDHVSARRQAREAFKDVQLGIDHILFKTQCDGLK 3
>BF635140 similar to PIR|G71415|G71 hypothetical protein - Arabidopsis
thaliana, partial (0%)
Length = 684
Score = 45.4 bits (106), Expect = 3e-05
Identities = 40/179 (22%), Positives = 77/179 (42%), Gaps = 1/179 (0%)
Frame = +3
Query: 149 IKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIG-VAKVLPKTK 207
+ G SMGG A+ + +AW G L++P + + + + G V +V P +
Sbjct: 51 LAGLSMGGMTAILGAIRDQSAWQGIILLSPAVDCPRNF--QLKIMEAIQGIVLRVAPNAR 224
Query: 208 LVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLL 267
LVPQ + + + +++LA + F K R+ L A + Q +++LP+L
Sbjct: 225 LVPQ-PRIPDITLNEVERKKLAE-DEFFDPKKVRVRMGNTFLAAWTHIAQNEHKLTLPIL 398
Query: 268 VMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWL 326
+ + D + P A++ + KD + + H L E + ++SW+
Sbjct: 399 IHYSTIDKVVYPPATERFAKNVASKDITVKPFDKFAHDLYMDTCKEESW---PQVVSWI 566
>BQ144020 homologue to PIR|B84620|B846 hypothetical protein At2g23070
[imported] - Arabidopsis thaliana, partial (14%)
Length = 573
Score = 36.2 bits (82), Expect = 0.018
Identities = 16/26 (61%), Positives = 18/26 (68%)
Frame = -1
Query: 309 GEPDETIFHVLDDIISWLDDHSSTKN 334
G DETI +L DIISWLD+HS N
Sbjct: 240 GTRDETITQILGDIISWLDEHSLKHN 163
>TC90757 similar to GP|17528996|gb|AAL38708.1 unknown protein {Arabidopsis
thaliana}, partial (41%)
Length = 717
Score = 36.2 bits (82), Expect = 0.018
Identities = 24/74 (32%), Positives = 40/74 (53%), Gaps = 2/74 (2%)
Frame = +3
Query: 83 KGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLH-GYIPSF-ENLVN 140
+G + + HG A T +F + V +L +GF FA D+ GFG SD GY ++ E +
Sbjct: 393 RGTIVFLHG-APTQSFSYRVVMSELGDAGFHCFAPDWIGFGFSDKPQPGYGFNYTEKEFH 569
Query: 141 DVIEHFSKIKGESM 154
D ++ ++ G S+
Sbjct: 570 DALDKLLEVWGSSL 611
>TC90355 similar to GP|9294510|dbj|BAB02772.1 gene_id:MDB19.2~unknown
protein {Arabidopsis thaliana}, partial (25%)
Length = 556
Score = 35.4 bits (80), Expect = 0.031
Identities = 28/86 (32%), Positives = 39/86 (44%), Gaps = 4/86 (4%)
Frame = +2
Query: 48 FQLPADGVKMEEIYEVNSRGLKVFSKSWIP----EKSPMKGIVYYCHGYADTCTFYFEGV 103
F L + +++ NSRG + +IP E P+ ++Y CHG + C
Sbjct: 173 FLLRGKWYQRKDVELKNSRGDALQCSHYIPIGSAEGKPLPCVIY-CHGNSG-CRADASEA 346
Query: 104 ARKLASSGFGVFALDYPGFGLSDGLH 129
A L S VF LD+ G GLS G H
Sbjct: 347 AIILLPSNITVFTLDFSGSGLSGGEH 424
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,105,185
Number of Sequences: 36976
Number of extensions: 135755
Number of successful extensions: 629
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 605
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 607
length of query: 336
length of database: 9,014,727
effective HSP length: 97
effective length of query: 239
effective length of database: 5,428,055
effective search space: 1297305145
effective search space used: 1297305145
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146563.12


