

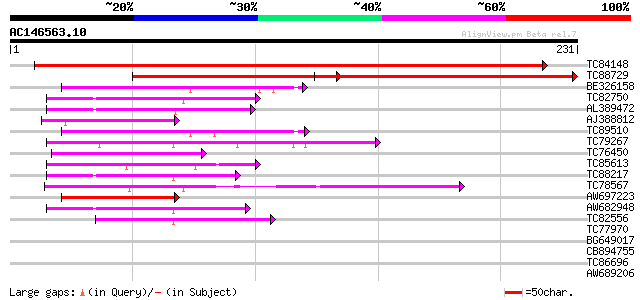
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.10 + phase: 0
(231 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unk... 344 2e-95
TC88729 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unk... 204 2e-53
BE326158 similar to GP|10177757|d phosphoribosylanthranilate tra... 57 6e-09
TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsi... 51 3e-07
AL389472 similar to GP|6648206|gb|A putative GTPase activating p... 49 1e-06
AJ388812 weakly similar to GP|15810203|g AT5g50170/K6A12_3 {Arab... 49 1e-06
TC89510 weakly similar to GP|22831331|dbj|BAC16176. contains EST... 49 2e-06
TC79267 similar to PIR|T00782|T00782 probable anthranilate phosp... 49 2e-06
TC76450 similar to GP|17065130|gb|AAL32719.1 putative protein {A... 48 3e-06
TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta ... 47 5e-06
TC88217 weakly similar to GP|18377801|gb|AAL67050.1 unknown prot... 46 1e-05
TC78567 similar to PIR|T04143|T04143 CLB1 protein - tomato, part... 44 4e-05
AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate tr... 43 9e-05
AW682948 weakly similar to PIR|E96525|E96 protein T1N15.21 [impo... 43 1e-04
TC82556 similar to GP|21536965|gb|AAM61306.1 putative zinc finge... 42 2e-04
TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {... 40 0.001
BG649017 similar to GP|9757890|db phosphoribosylanthranilate tra... 35 0.024
CB894755 similar to GP|9757890|db phosphoribosylanthranilate tra... 35 0.024
TC86696 similar to GP|3860331|emb|CAA10133.1 hypothetical protei... 35 0.032
AW689206 similar to GP|17473880|gb| putative protein {Arabidopsi... 30 0.78
>TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unknown
protein {Arabidopsis thaliana}, partial (16%)
Length = 840
Score = 344 bits (882), Expect = 2e-95
Identities = 171/209 (81%), Positives = 187/209 (88%)
Frame = +1
Query: 11 SGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP 70
SG NGWIELVLIE RDL+AADLRGTSDP+VRV+YGN KKRTKV++KT+ P+W+QTLEF
Sbjct: 97 SGVTNGWIELVLIEARDLIAADLRGTSDPFVRVNYGNLKKRTKVVHKTINPRWDQTLEFL 276
Query: 71 DDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKV 130
DDGSPL L+VKDHNALLPTSSIGECVVEYQ LPPNQ +DKWIPLQGVK GEIHIQI RKV
Sbjct: 277 DDGSPLTLHVKDHNALLPTSSIGECVVEYQSLPPNQTSDKWIPLQGVKSGEIHIQIARKV 456
Query: 131 PEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLEDTQEG 190
PE+Q RQS D EPSL+KLHQ P+QIK+M K R IEDGNLE LSTTLSELETLEDTQEG
Sbjct: 457 PEIQTRQSPDFEPSLTKLHQSPSQIKEMTKKVRYLIEDGNLEELSTTLSELETLEDTQEG 636
Query: 191 YVAQLETEQMLLLSKIKELGQEIINSSPS 219
Y+AQLETEQMLL+SKI ELGQEIINSSPS
Sbjct: 637 YIAQLETEQMLLISKINELGQEIINSSPS 723
Score = 26.6 bits (57), Expect = 8.6
Identities = 9/16 (56%), Positives = 13/16 (81%)
Frame = +1
Query: 111 WIPLQGVKRGEIHIQI 126
WIPL+GV GE+ ++I
Sbjct: 4 WIPLEGVSSGELRLKI 51
>TC88729 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1047
Score = 204 bits (520), Expect = 2e-53
Identities = 107/107 (100%), Positives = 107/107 (100%)
Frame = +3
Query: 125 QITRKVPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETL 184
QITRKVPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETL
Sbjct: 222 QITRKVPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETL 401
Query: 185 EDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLSRRISESVN 231
EDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLSRRISESVN
Sbjct: 402 EDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLSRRISESVN 542
Score = 162 bits (409), Expect = 1e-40
Identities = 75/85 (88%), Positives = 80/85 (93%)
Frame = +1
Query: 51 RTKVIYKTLTPQWNQTLEFPDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADK 110
RTKVIYKTLTPQWNQTLEFPDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADK
Sbjct: 1 RTKVIYKTLTPQWNQTLEFPDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADK 180
Query: 111 WIPLQGVKRGEIHIQITRKVPEMQK 135
WIPLQGVKRGEIHI++ K + ++
Sbjct: 181 WIPLQGVKRGEIHIKLLEKFQKCRR 255
>BE326158 similar to GP|10177757|d phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(15%)
Length = 618
Score = 57.0 bits (136), Expect = 6e-09
Identities = 37/108 (34%), Positives = 54/108 (49%), Gaps = 8/108 (7%)
Frame = +3
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---GSPLML 78
+++ ++L L T DPYV V GN+K RTK + K P+WNQ F D S L +
Sbjct: 189 VVKAKNLTLNSLTSTCDPYVEVRLGNYKGRTKHLDKRSNPEWNQVYAFSKDQIQSSILEV 368
Query: 79 YVKDHNALLPTSSIGECVVEYQ----RLPPNQ-MADKWIPLQGVKRGE 121
VKD + IG + R+PP+ +A +W L+ +RGE
Sbjct: 369 IVKDKETVGRDDYIGRVAFDLNEVPTRVPPDSPLAPQWYRLED-RRGE 509
>TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsis
thaliana}, partial (95%)
Length = 623
Score = 51.2 bits (121), Expect = 3e-07
Identities = 31/89 (34%), Positives = 47/89 (51%), Gaps = 2/89 (2%)
Frame = +3
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF--PDDG 73
G +++++++G+ LV D + TSDPYV + GN +TKVI L P WN+ L F +
Sbjct: 78 GQLKVIVVQGKRLVIRDFK-TSDPYVVLKLGNQTAKTKVINSCLNPVWNEELNFTLTEPL 254
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRL 102
L L V D + L +G + Q L
Sbjct: 255 GVLNLEVFDKDLLKADDKMGNAFINLQPL 341
>AL389472 similar to GP|6648206|gb|A putative GTPase activating protein
{Arabidopsis thaliana}, partial (24%)
Length = 487
Score = 49.3 bits (116), Expect = 1e-06
Identities = 28/87 (32%), Positives = 49/87 (56%), Gaps = 2/87 (2%)
Frame = +1
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQT--LEFPDDG 73
G I++ +++G +L D+ TSDPYV + G+ +T+VI L P WN++ L P++
Sbjct: 229 GLIKVNVVKGTNLAIRDIV-TSDPYVILSLGHQSVKTRVIKNNLNPVWNESLMLSIPENI 405
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQ 100
PL + V D ++ +GE ++ Q
Sbjct: 406 PPLKIIVYDKDSFKNDDFMGEAEIDIQ 486
>AJ388812 weakly similar to GP|15810203|g AT5g50170/K6A12_3 {Arabidopsis
thaliana}, partial (14%)
Length = 673
Score = 49.3 bits (116), Expect = 1e-06
Identities = 26/57 (45%), Positives = 34/57 (59%), Gaps = 1/57 (1%)
Frame = +2
Query: 14 GNGWIELV-LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G+GW+ V LIEG DLV+ + G SDPYV + + V +T PQWN+ LEF
Sbjct: 272 GDGWVVTVALIEGVDLVSLESTGLSDPYVVFTCNGQTRSSSVKLETSDPQWNEILEF 442
>TC89510 weakly similar to GP|22831331|dbj|BAC16176. contains ESTs
AU165964(S1723) D40008(S1723)~phosphoribosylanthranilate
transferase-like protein, partial (21%)
Length = 1195
Score = 48.9 bits (115), Expect = 2e-06
Identities = 33/108 (30%), Positives = 52/108 (47%), Gaps = 7/108 (6%)
Frame = +3
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---GSPLML 78
+++ RDL DL G+ DPYV V GNFK T K +P+WN F + + L +
Sbjct: 846 VVKARDLPRMDLTGSLDPYVIVKVGNFKGTTNHFEKNNSPEWNLVFAFAKENQQATTLEV 1025
Query: 79 YVKD----HNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEI 122
+KD H+ + T V + P + +A +W + K+GE+
Sbjct: 1026VIKDKDTIHDDFVGTVRFDLYDVPKRVPPDSPLAPQWYRIXN-KKGEM 1166
Score = 35.4 bits (80), Expect = 0.019
Identities = 17/48 (35%), Positives = 26/48 (53%)
Frame = +3
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
++ DLVA D G+S +V + + + K RT K L+P WN+ F
Sbjct: 36 VVGAHDLVAKDGEGSSTTFVELEFDDQKFRTTTKDKDLSPYWNEIFYF 179
>TC79267 similar to PIR|T00782|T00782 probable anthranilate
phosphoribosyltransferase (EC 2.4.2.18) T22J18.21 -
Arabidopsis thaliana, partial (60%)
Length = 1612
Score = 48.5 bits (114), Expect = 2e-06
Identities = 39/151 (25%), Positives = 72/151 (46%), Gaps = 15/151 (9%)
Frame = +3
Query: 16 GWIELVLIEGRDLVAADLRG--TSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPD 71
G++EL ++ R+L+ + T+D Y YGN RT+ + TL+P+WN+ T E D
Sbjct: 204 GYLELGILSARNLLPMKGKDGRTTDAYCVAKYGNKWVRTRTLLDTLSPRWNEQYTWEVHD 383
Query: 72 DGSPLMLYVKDHNALLPTSS-----IGECVVEYQRLPPNQMADKWIPL-----QGVKR-G 120
+ + + V D++ L +S IG+ + L +++ + PL G+K+ G
Sbjct: 384 PCTVITVSVFDNHHLNGSSDHKDQRIGKVRIRLSTLETDRVYTHYYPLLVLQPNGLKKNG 563
Query: 121 EIHIQITRKVPEMQKRQSMDSEPSLSKLHQI 151
E+H+ + + P L K+H +
Sbjct: 564 ELHLAVRFTCTAWVNMVAQYGRPLLPKMHYV 656
>TC76450 similar to GP|17065130|gb|AAL32719.1 putative protein {Arabidopsis
thaliana}, partial (83%)
Length = 2171
Score = 48.1 bits (113), Expect = 3e-06
Identities = 22/63 (34%), Positives = 37/63 (57%)
Frame = +1
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDGSPLM 77
I+L L+ ++L+AA+L GTSDPY + GN K+ + ++ + P W + F D P+
Sbjct: 484 IKLELLAAKNLIAANLNGTSDPYTIITCGNEKRFSSMVPGSRNPMWGEEFNFSVDELPVQ 663
Query: 78 LYV 80
+ V
Sbjct: 664 INV 672
>TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta {Homo
sapiens}, partial (3%)
Length = 584
Score = 47.4 bits (111), Expect = 5e-06
Identities = 29/89 (32%), Positives = 50/89 (55%), Gaps = 2/89 (2%)
Frame = +1
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYG-NFKKRTKVIYKTLTPQWNQTLEFPDDGS 74
G ++++++ G+ L D G SDPYV + ++K+RT T+ P+WN+T F DG
Sbjct: 85 GTLKVIVVRGKKLKDEDAIGKSDPYVELWIKKDYKQRTTTKKGTVNPEWNETFTFNVDGD 264
Query: 75 -PLMLYVKDHNALLPTSSIGECVVEYQRL 102
L L V D + ++ IGE V+ +++
Sbjct: 265 HHLYLKVLDDD-VVSDDKIGEAKVDLKQV 348
>TC88217 weakly similar to GP|18377801|gb|AAL67050.1 unknown protein
{Arabidopsis thaliana}, partial (94%)
Length = 857
Score = 46.2 bits (108), Expect = 1e-05
Identities = 26/81 (32%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Frame = +3
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G +++ + G +L D+ +SDPYV + K +T+V+ K L P+WN+ TL D
Sbjct: 96 GLLKIHVQRGVNLAIRDVV-SSDPYVVIKMAKQKLKTRVVKKNLNPEWNEDLTLSISDPH 272
Query: 74 SPLMLYVKDHNALLPTSSIGE 94
+P+ LYV D + +G+
Sbjct: 273 TPIHLYVYDKDTFSLDDKMGD 335
>TC78567 similar to PIR|T04143|T04143 CLB1 protein - tomato, partial (53%)
Length = 1159
Score = 44.3 bits (103), Expect = 4e-05
Identities = 44/175 (25%), Positives = 78/175 (44%), Gaps = 4/175 (2%)
Frame = +3
Query: 15 NGWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-FKKRTKVIYKTLTPQWNQTLEF---P 70
+G +++ +++ DL ++ G SDPYV ++ FK +TKVI L P W+QT E
Sbjct: 171 HGSLKVTIVKATDLKNMEMIGKSDPYVVLYIRPLFKVKTKVINNNLNPVWDQTFELIAED 350
Query: 71 DDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKV 130
+ L+L V D + IG+ DK + + + E+ +Q T K
Sbjct: 351 KETQSLILEVFDED-------IGQ--------------DKRLGIVKLPLIELEVQ-TEKE 464
Query: 131 PEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLE 185
E++ S+D+ K + +K + +F + + LE L E + L+
Sbjct: 465 LELRLLSSLDTLKVKDKKDRGTLTVKVLYYQFNKEEQLAALEAEKAILEERKKLK 629
>AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(5%)
Length = 778
Score = 43.1 bits (100), Expect = 9e-05
Identities = 16/48 (33%), Positives = 30/48 (62%)
Frame = +3
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
+I+ ++L D GTS PY+ + + +++T+ + + L P WN+TL F
Sbjct: 168 VIDAQNLAPKDGHGTSSPYIVIDFHGQRRKTRTLVRDLNPVWNETLSF 311
>AW682948 weakly similar to PIR|E96525|E96 protein T1N15.21 [imported] -
Arabidopsis thaliana, partial (72%)
Length = 576
Score = 42.7 bits (99), Expect = 1e-04
Identities = 28/85 (32%), Positives = 42/85 (48%), Gaps = 2/85 (2%)
Frame = +3
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G I L + +G +L+ D R TSDPYV V +T V+ P+WN+ TL D
Sbjct: 21 GLIRLRIKKGTNLIPHDSR-TSDPYVLVTMEEQTLKTAVVNDNCHPEWNEELTLYIKDVN 197
Query: 74 SPLMLYVKDHNALLPTSSIGECVVE 98
+P+ L V D + +GE ++
Sbjct: 198 TPIHLIVCDKDTFTVDDKMGEADID 272
>TC82556 similar to GP|21536965|gb|AAM61306.1 putative zinc finger and C2
domain protein {Arabidopsis thaliana}, partial (40%)
Length = 756
Score = 42.4 bits (98), Expect = 2e-04
Identities = 25/75 (33%), Positives = 37/75 (49%), Gaps = 2/75 (2%)
Frame = +3
Query: 36 TSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDGSPLMLYVKDHNALLPTSSIG 93
+SDPYV ++ G +T V+ L P WN+ L P+ L L V DH+ +G
Sbjct: 39 SSDPYVVLNLGTQTVQTSVMRSNLNPVWNEEHMLSVPEHYGQLKLKVFDHDTFSADDIMG 218
Query: 94 ECVVEYQRLPPNQMA 108
E ++ Q L + MA
Sbjct: 219 EADIDLQSLITSAMA 263
>TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {Arabidopsis
thaliana}, partial (97%)
Length = 2062
Score = 39.7 bits (91), Expect = 0.001
Identities = 37/149 (24%), Positives = 65/149 (42%), Gaps = 17/149 (11%)
Frame = +2
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFK---KRTKVIYKTLTPQWNQTLEF--- 69
G + + ++ L DL G SDPYV++ + K K+T V +K L P+WN+
Sbjct: 950 GILHVKVLHAMKLKKKDLLGASDPYVKLKLTDDKMPSKKTTVKHKNLNPEWNEEFNLVVK 1129
Query: 70 PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL---------QGVK-R 119
+ L L V D + +G V+ + + P + + L Q K R
Sbjct: 1130 DPETQVLQLNVYDWEQVGKHDKMGMNVITLKEVSPEEPKRFTLDLLKTMDPNDAQNEKSR 1309
Query: 120 GEIHIQITRK-VPEMQKRQSMDSEPSLSK 147
G+I +++T K + E + + D ++ K
Sbjct: 1310 GQIVVEVTYKPLNEEEMGKGFDETQTIPK 1396
>BG649017 similar to GP|9757890|db phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(15%)
Length = 554
Score = 35.0 bits (79), Expect = 0.024
Identities = 43/164 (26%), Positives = 71/164 (43%), Gaps = 28/164 (17%)
Frame = +3
Query: 16 GWIELVLIEGRDLV---AADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFP 70
G IEL +I ++L+ + + ++D Y YGN RT+ + L P+WN+ T +
Sbjct: 42 GTIELGIIGCKNLIPMKTVNGKSSTDGYCVAKYGNKWVRTRTVSDNLEPKWNEQYTWKVF 221
Query: 71 DDGSPLMLYVKD---------------HNALLPTSSIGECVVEYQRLPPNQMADKWIPL- 114
D + L + V D + + P IG+ + L ++ PL
Sbjct: 222 DPSTVLTIGVFDSFSVFESDNSKTEMTNESTRPDFRIGKVRIRISTLQTGRVYKNTYPLL 401
Query: 115 ----QGVKR-GEIHIQITRKVPEMQKRQSMD--SEPSLSKLHQI 151
G+K+ GEI I I R V +Q+ + S+P L +H I
Sbjct: 402 VLTHGGLKKMGEIEIAI-RFVRTVQRLDFLHVYSQPMLPLMHHI 530
>CB894755 similar to GP|9757890|db phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana},
partial (10%)
Length = 674
Score = 35.0 bits (79), Expect = 0.024
Identities = 14/38 (36%), Positives = 23/38 (59%)
Frame = +3
Query: 32 DLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
D GTS Y+ V + +++T+ + + L P WN+TL F
Sbjct: 6 DGHGTSSLYIVVDFYGQRRKTRTLVRDLNPVWNETLSF 119
>TC86696 similar to GP|3860331|emb|CAA10133.1 hypothetical protein {Cicer
arietinum}, partial (90%)
Length = 733
Score = 34.7 bits (78), Expect = 0.032
Identities = 25/85 (29%), Positives = 38/85 (44%), Gaps = 3/85 (3%)
Frame = +1
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHY-GNFKKRTKVIYKTLTPQWNQTLEF--PDD 72
G +E+VLI + L D + DPYV + Y K T PQWN+T F D
Sbjct: 94 GTLEVVLISAKGLEDNDFLSSIDPYVILTYRAQEHKSTVQEGAGSNPQWNETFLFTVSDT 273
Query: 73 GSPLMLYVKDHNALLPTSSIGECVV 97
L L + + + ++GE ++
Sbjct: 274 AYELNLKIMEKDNYSADDNLGEVII 348
>AW689206 similar to GP|17473880|gb| putative protein {Arabidopsis thaliana},
partial (6%)
Length = 615
Score = 30.0 bits (66), Expect = 0.78
Identities = 20/53 (37%), Positives = 27/53 (50%)
Frame = +2
Query: 174 LSTTLSELETLEDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLSRRI 226
L T SELE+ T E A E+E+ LL +++EL +INS S I
Sbjct: 329 LETQKSELESRRITLEKREAHNESERRKLLEEMEELNLWMINSGKSQKTGNMI 487
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,398,398
Number of Sequences: 36976
Number of extensions: 80038
Number of successful extensions: 348
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 345
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 348
length of query: 231
length of database: 9,014,727
effective HSP length: 93
effective length of query: 138
effective length of database: 5,575,959
effective search space: 769482342
effective search space used: 769482342
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146563.10


