

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146561.3 + phase: 0
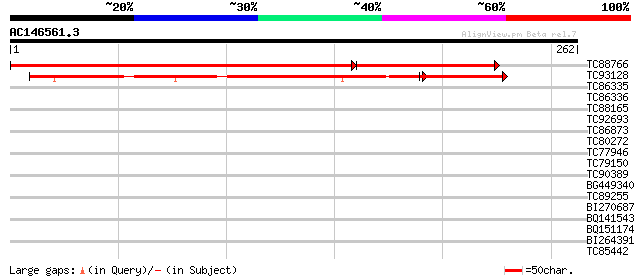
(262 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88766 similar to PIR|T04745|T04745 hypothetical protein F6G17.... 321 e-122
TC93128 similar to GP|20160696|dbj|BAB89639. hypothetical protei... 184 2e-62
TC86335 similar to GP|14626761|gb|AAK71662.1 mature anther-speci... 33 0.15
TC86336 similar to GP|14626761|gb|AAK71662.1 mature anther-speci... 33 0.15
TC88165 similar to GP|7716952|gb|AAF68626.1| NAC1 {Medicago trun... 30 0.94
TC92693 similar to PIR|T48328|T48328 importin alpha-like protein... 30 0.94
TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 30 1.2
TC80272 similar to GP|15620810|dbj|BAB67768. RPS6-like protein {... 29 1.6
TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing... 29 1.6
TC79150 similar to GP|18461304|dbj|BAB84499. putative receptor-l... 29 1.6
TC90389 similar to GP|21592590|gb|AAM64539.1 unknown {Arabidopsi... 28 2.7
BG449340 similar to PIR|B84799|B847 hypothetical protein At2g379... 28 3.6
TC89255 similar to GP|3135257|gb|AAC16457.1| unknown protein {Ar... 28 4.7
BI270687 weakly similar to GP|1903347|gb EST gb|ATTS5672 comes f... 28 4.7
BQ141543 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 27 6.1
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 27 8.0
BI264391 similar to GP|20161425|dbj hypothetical protein {Oryza ... 27 8.0
TC85442 similar to GP|13543783|gb|AAH06040.1 Unknown (protein fo... 27 8.0
>TC88766 similar to PIR|T04745|T04745 hypothetical protein F6G17.160 -
Arabidopsis thaliana, partial (34%)
Length = 813
Score = 321 bits (822), Expect(2) = e-122
Identities = 160/160 (100%), Positives = 160/160 (100%)
Frame = +3
Query: 1 MELSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNP 60
MELSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNP
Sbjct: 135 MELSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNP 314
Query: 61 QRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPS 120
QRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPS
Sbjct: 315 QRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPS 494
Query: 121 LRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAKQ 160
LRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAKQ
Sbjct: 495 LRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAKQ 614
Score = 136 bits (342), Expect(2) = e-122
Identities = 65/66 (98%), Positives = 65/66 (98%)
Frame = +1
Query: 161 FDTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRER 220
FDTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRY RELPGPMRER
Sbjct: 616 FDTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYSRELPGPMRER 795
Query: 221 VYGLIG 226
VYGLIG
Sbjct: 796 VYGLIG 813
>TC93128 similar to GP|20160696|dbj|BAB89639. hypothetical protein~similar
to Arabidopsis thaliana chromosome 4 At4g37510, partial
(23%)
Length = 673
Score = 184 bits (467), Expect(2) = 2e-62
Identities = 104/189 (55%), Positives = 127/189 (67%), Gaps = 5/189 (2%)
Frame = +2
Query: 10 SPKPTNFSFS--SSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLLKEL 67
SPKP +FSFS +S S PIQ T + LH+ A+ +P N+PQRLLKEL
Sbjct: 14 SPKPHSFSFSFSTSISQNPIQFPTNKTITPQKTLHIYALTTNPLPP----NSPQRLLKEL 181
Query: 68 AERKKITN-PKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPSLRLNNA 126
+ K T+ P K+PPR ILKPPL +K L +NSPQL+L +FPLLSSCLP L
Sbjct: 182 EQHKNKTHSPNKKTPPRTTILKPPLRNKNL----INSPQLTLSTFPLLSSCLPHAPLKTT 349
Query: 127 DKLWIDEYLLEAKQALGYSLEPSETL--EDDNPAKQFDTLLYLAFQHPSCERTKQKHVRS 184
D W++ +LLE KQALGY L+P + + E+DNPAKQFDTLLYLAFQHP+C R K +HVRS
Sbjct: 350 DSAWMETHLLEVKQALGYPLQPFDEMLGEEDNPAKQFDTLLYLAFQHPTC-RGKGRHVRS 526
Query: 185 GHSRLFFLG 193
HSRLFF G
Sbjct: 527 AHSRLFFFG 553
Score = 72.0 bits (175), Expect(2) = 2e-62
Identities = 32/41 (78%), Positives = 37/41 (90%)
Frame = +3
Query: 190 FFLGQYVLELALAEFFLQRYPRELPGPMRERVYGLIGKENL 230
FFLGQYV+ELAL EFFLQRYPRE GPMRERV+GL+GK ++
Sbjct: 543 FFLGQYVIELALGEFFLQRYPREAVGPMRERVFGLVGKHSV 665
>TC86335 similar to GP|14626761|gb|AAK71662.1 mature anther-specific protein
LAT61 {Lycopersicon esculentum}, partial (60%)
Length = 1595
Score = 32.7 bits (73), Expect = 0.15
Identities = 32/118 (27%), Positives = 49/118 (41%), Gaps = 10/118 (8%)
Frame = +1
Query: 13 PTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLLKELAERKK 72
PTN S SS P P + +S ++P + + PT+ NP L L +
Sbjct: 427 PTNMSACSSMQPSPQSSSFPSPQSSSFPSPIPSYPTSPTR-MDGITNPSSFL--LPFIRN 597
Query: 73 ITNPKSKSPPRRYI----LKPPLDDKKLAERFLNSPQLSLKSF------PLLSSCLPS 120
IT+ + PP R + PPL + ++R + L SF PL ++ PS
Sbjct: 598 ITSIPTNLPPLRISNSAPVTPPLSSPRSSKRKADFESLCNGSFNSSFRHPLFATSAPS 771
>TC86336 similar to GP|14626761|gb|AAK71662.1 mature anther-specific protein
LAT61 {Lycopersicon esculentum}, partial (37%)
Length = 778
Score = 32.7 bits (73), Expect = 0.15
Identities = 32/118 (27%), Positives = 49/118 (41%), Gaps = 10/118 (8%)
Frame = +1
Query: 13 PTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLLKELAERKK 72
PTN S SS P P + +S ++P + + PT+ NP L L +
Sbjct: 307 PTNMSACSSMQPSPQSSSFPSPQSSSFPSPIPSYPTSPTR-MDGITNPSSFL--LPFIRN 477
Query: 73 ITNPKSKSPPRRYI----LKPPLDDKKLAERFLNSPQLSLKSF------PLLSSCLPS 120
IT+ + PP R + PPL + ++R + L SF PL ++ PS
Sbjct: 478 ITSIPTNLPPLRISNSAPVTPPLSSPRSSKRKADFESLCNGSFNSSFRHPLFATSAPS 651
>TC88165 similar to GP|7716952|gb|AAF68626.1| NAC1 {Medicago truncatula},
partial (61%)
Length = 1231
Score = 30.0 bits (66), Expect = 0.94
Identities = 21/55 (38%), Positives = 31/55 (56%)
Frame = -2
Query: 3 LSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRN 57
L+SSF PKP +F SSF P+P IL+ N ++ PL + + I + PR+
Sbjct: 744 LTSSF---PKP---NFRSSFHPWPTSILLLN-RTEKVPLQIPVLEIR*FWEIPRS 601
>TC92693 similar to PIR|T48328|T48328 importin alpha-like protein -
Arabidopsis thaliana, partial (32%)
Length = 770
Score = 30.0 bits (66), Expect = 0.94
Identities = 21/62 (33%), Positives = 29/62 (45%), Gaps = 4/62 (6%)
Frame = -3
Query: 3 LSSSFS----LSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNN 58
L+SSFS LS S S+SF PFP I +K + P +++ D TK
Sbjct: 492 LTSSFS*N*NLSMASIPSSCSTSFGPFPFGIPLKTSSIN*SPSLAASISADLTKSIKPGK 313
Query: 59 NP 60
+P
Sbjct: 312 HP 307
>TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (53%)
Length = 1475
Score = 29.6 bits (65), Expect = 1.2
Identities = 34/111 (30%), Positives = 51/111 (45%), Gaps = 4/111 (3%)
Frame = -3
Query: 8 SLSPKPTNFSFS---SSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLL 64
SLSP PTN +FS S + P+ L T S + ++V I + FP + P R L
Sbjct: 603 SLSPSPTNLAFSFWPSLVTGIPVHRL---TTSAIWSGPTVSVNIKFSSVFP-SVLPSRFL 436
Query: 65 KELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQL-SLKSFPLL 114
++ +S + R Y L K+ + RF S ++ S+ SF LL
Sbjct: 435 IAVSTSSIAPYRRSAALARSYRLSASCSAKRSSSRFCFSLRISSILSFSLL 283
>TC80272 similar to GP|15620810|dbj|BAB67768. RPS6-like protein
{Arabidopsis thaliana}, partial (35%)
Length = 1026
Score = 29.3 bits (64), Expect = 1.6
Identities = 16/61 (26%), Positives = 29/61 (47%)
Frame = +3
Query: 12 KPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLLKELAERK 71
K +++ SS P I +L N N+P H L+ + ++ F NNP++ + +
Sbjct: 3 KGSSYLLPSSKPPKTITMLASNPLRFNFPNHHLSSSSSFSRQFSLQNNPRKSFSLIMNAR 182
Query: 72 K 72
K
Sbjct: 183K 185
>TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing factor
{Cicer arietinum}, complete
Length = 2061
Score = 29.3 bits (64), Expect = 1.6
Identities = 24/86 (27%), Positives = 40/86 (45%)
Frame = -3
Query: 3 LSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQR 62
LS SFSLS + SFS S S + + S ++ L L+ ++ + R+ +
Sbjct: 514 LSRSFSLSLSLLSLSFSLSLSLSLSLPPLSTSFSLSFSLLPLSPSLSR*RPEDRSRCSRS 335
Query: 63 LLKELAERKKITNPKSKSPPRRYILK 88
+ L+ + P S SPPR +L+
Sbjct: 334 RSRSLSLSRSFLEPFSSSPPRDLLLR 257
>TC79150 similar to GP|18461304|dbj|BAB84499. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(7%)
Length = 1077
Score = 29.3 bits (64), Expect = 1.6
Identities = 30/108 (27%), Positives = 48/108 (43%), Gaps = 7/108 (6%)
Frame = +3
Query: 12 KPTNFSFSSSFSPFPIQILIKNT-----KSRNYPLHVLAVAIDPTKDFPRNNNPQRLL-- 64
K T +SSFSPFP+Q+ ++N + N + +L + DFP + LL
Sbjct: 372 KRTPIWSTSSFSPFPLQLKLQNNGNLVLSTTNGNISILWQSF----DFPTDT----LLPG 527
Query: 65 KELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFP 112
+E+ ER + + KS++ K D+ SP LS +P
Sbjct: 528 QEINERATLVSSKSETNYSSGFYKFYFDNDNALRLLFKSPLLSSVYWP 671
>TC90389 similar to GP|21592590|gb|AAM64539.1 unknown {Arabidopsis
thaliana}, partial (22%)
Length = 658
Score = 28.5 bits (62), Expect = 2.7
Identities = 18/78 (23%), Positives = 34/78 (43%)
Frame = +1
Query: 100 FLNSPQLSLKSFPLLSSCLPSLRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAK 159
F +P+L L+ SS L N D + +++ + + L + + NPA
Sbjct: 145 FSPTPKLQLRLLRASSSSTTFLDTNPTDSVIVEKEITRSSNPLACPVCYNSLTWTTNPAL 324
Query: 160 QFDTLLYLAFQHPSCERT 177
DT+ + Q +C++T
Sbjct: 325 SIDTISGSSLQCSTCQKT 378
>BG449340 similar to PIR|B84799|B847 hypothetical protein At2g37950
[imported] - Arabidopsis thaliana, partial (22%)
Length = 627
Score = 28.1 bits (61), Expect = 3.6
Identities = 14/37 (37%), Positives = 21/37 (55%)
Frame = -2
Query: 66 ELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLN 102
E+ E+ N S PP R PP+++++L RFLN
Sbjct: 467 EMTEQWSEANDASVXPPTRR--SPPVENQRLLHRFLN 363
>TC89255 similar to GP|3135257|gb|AAC16457.1| unknown protein {Arabidopsis
thaliana}, partial (28%)
Length = 1420
Score = 27.7 bits (60), Expect = 4.7
Identities = 24/92 (26%), Positives = 37/92 (40%), Gaps = 4/92 (4%)
Frame = +3
Query: 4 SSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRL 63
SS FS+ P+P + S SSS L ++ P K P ++ P
Sbjct: 174 SSLFSILPQPKSSSSSSS----------------------LFNSLPPPKQQPSSDTPIDA 287
Query: 64 LKELAERKKITNPKS---KSPPRRYI-LKPPL 91
+ + PKS + PP+R + KPP+
Sbjct: 288 PSNFTQISSLPKPKSQIHQQPPKRVVQFKPPI 383
>BI270687 weakly similar to GP|1903347|gb EST gb|ATTS5672 comes from this
gene. {Arabidopsis thaliana}, partial (17%)
Length = 665
Score = 27.7 bits (60), Expect = 4.7
Identities = 23/90 (25%), Positives = 42/90 (46%), Gaps = 3/90 (3%)
Frame = +3
Query: 68 AERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPSLRLNNAD 127
AE K + ++K ++L + + LA + + LK + +++ L +L LNN
Sbjct: 66 AENAKTADSETKKDISEFMLHLAQEAEDLASKERQNYSPILKKWNAIAAALAALTLNNCY 245
Query: 128 KLWIDEYLLEAKQ---ALGYSLEPSETLED 154
+ +YL E K L L+ ++ LED
Sbjct: 246 GHVLKQYLSEIKSITVELIIVLQKAKRLED 335
>BQ141543 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (15%)
Length = 1271
Score = 27.3 bits (59), Expect = 6.1
Identities = 26/88 (29%), Positives = 36/88 (40%), Gaps = 10/88 (11%)
Frame = +1
Query: 2 ELSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYP---LHVLAVAIDPTKDFPRN- 57
E + F P F+F SSF +P I IK + +P H P+ F RN
Sbjct: 109 ETDTGFLEGPHGPYFNFKSSFRHYPPPIKIKPIQPTPHPHRKFHQKQHQNSPSSQFYRNQ 288
Query: 58 ------NNPQRLLKELAERKKITNPKSK 79
N +RL+ A R IT P ++
Sbjct: 289 PQHNDDNRKKRLVDHQAPR--ITIPGAR 366
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 26.9 bits (58), Expect = 8.0
Identities = 13/41 (31%), Positives = 19/41 (45%)
Frame = +2
Query: 50 PTKDFPRNNNPQRLLKELAERKKITNPKSKSPPRRYILKPP 90
P + P PQ+ KE +++K S PPR +PP
Sbjct: 56 PKPETPNRKKPQKNKKEPPKKRKTPPEASHPPPRPRPPRPP 178
>BI264391 similar to GP|20161425|dbj hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (13%)
Length = 634
Score = 26.9 bits (58), Expect = 8.0
Identities = 21/84 (25%), Positives = 34/84 (40%), Gaps = 11/84 (13%)
Frame = -3
Query: 16 FSFSSSFSPFPIQIL-----------IKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLL 64
FSFS SFSP P Q L + ++ PL + +A ++ P
Sbjct: 620 FSFSFSFSPLPPQSLEXFASLTSEDQFASPLPKSSPLESVPIATGKSEQVP*AAEAPTEQ 441
Query: 65 KELAERKKITNPKSKSPPRRYILK 88
+ L + + P++ PRR L+
Sbjct: 440 QRLVALRSLLRPETDLEPRRRRLR 369
>TC85442 similar to GP|13543783|gb|AAH06040.1 Unknown (protein for MGC:7642)
{Mus musculus}, partial (62%)
Length = 585
Score = 26.9 bits (58), Expect = 8.0
Identities = 14/18 (77%), Positives = 14/18 (77%)
Frame = -1
Query: 106 LSLKSFPLLSSCLPSLRL 123
L L SFPLLSS LPSL L
Sbjct: 174 LVLPSFPLLSSLLPSLLL 121
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,980,590
Number of Sequences: 36976
Number of extensions: 146831
Number of successful extensions: 1060
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 1047
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1054
length of query: 262
length of database: 9,014,727
effective HSP length: 94
effective length of query: 168
effective length of database: 5,538,983
effective search space: 930549144
effective search space used: 930549144
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146561.3


