

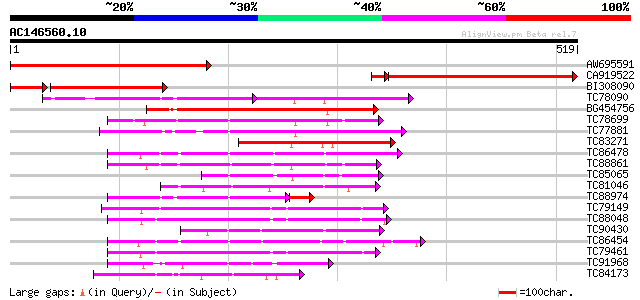
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146560.10 + phase: 0
(519 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW695591 similar to PIR|T51338|T51 mitogen-activated protein kin... 371 e-103
CA919522 similar to PIR|S53804|S53 protein kinase NPK2 (EC 2.7.1... 356 e-100
BI308090 similar to GP|21655213|gb mitogen-activated protein kin... 217 9e-72
TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sa... 129 2e-59
BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabac... 200 1e-51
TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase ... 175 4e-44
TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase ki... 161 5e-40
TC83271 similar to GP|15528441|emb|CAC69138. MAP kinase kinase {... 155 5e-38
TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 prote... 140 2e-33
TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative pro... 117 1e-26
TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated p... 113 2e-25
TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein ki... 111 6e-25
TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Bra... 96 3e-22
TC79149 similar to PIR|T46059|T46059 MAP kinase - Arabidopsis th... 102 5e-22
TC88048 similar to GP|20302596|dbj|BAB91125. Ser/Thr kinase {Ara... 100 1e-21
TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 98 7e-21
TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine... 94 1e-19
TC79461 similar to GP|9758254|dbj|BAB08753.1 contains similarity... 93 3e-19
TC91968 similar to PIR|T02550|T02550 NPK1-related protein kinase... 92 5e-19
TC84173 similar to PIR|A71407|A71407 probable Ste20-like kinase ... 91 8e-19
>AW695591 similar to PIR|T51338|T51 mitogen-activated protein kinase kinase
(EC 2.7.1.-) 3 [imported] - Arabidopsis thaliana,
partial (34%)
Length = 649
Score = 371 bits (953), Expect = e-103
Identities = 184/184 (100%), Positives = 184/184 (100%)
Frame = +2
Query: 1 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYTFSDGGTVNLLSRSYGVYNINELGLQK 60
MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYTFSDGGTVNLLSRSYGVYNINELGLQK
Sbjct: 98 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYTFSDGGTVNLLSRSYGVYNINELGLQK 277
Query: 61 CSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEK 120
CSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEK
Sbjct: 278 CSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEK 457
Query: 121 EKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP 180
EKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP
Sbjct: 458 EKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP 637
Query: 181 EPIL 184
EPIL
Sbjct: 638 EPIL 649
>CA919522 similar to PIR|S53804|S53 protein kinase NPK2 (EC 2.7.1.-) - common
tobacco, partial (33%)
Length = 799
Score = 356 bits (913), Expect(2) = e-100
Identities = 171/173 (98%), Positives = 171/173 (98%)
Frame = -1
Query: 347 KVDLPGFVRSVFDPTQRMKDLADMLTIHYYLLFDGPDDSWQHTRNFYNENSIFSFSGKQH 406
KVDLPG RSVFDPTQRMKDLADMLTIHYYLLFDGPDDSWQHTRNFYNENSIFSFSGKQH
Sbjct: 751 KVDLPGLFRSVFDPTQRMKDLADMLTIHYYLLFDGPDDSWQHTRNFYNENSIFSFSGKQH 572
Query: 407 IGPNNIFTTLSSIRTTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLIC 466
IGPNNIFTTLSSIRTTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLIC
Sbjct: 571 IGPNNIFTTLSSIRTTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLIC 392
Query: 467 GDGVQVEGLPNFKDLGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYINQ 519
GDGVQVEGLPNFKDLGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYINQ
Sbjct: 391 GDGVQVEGLPNFKDLGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYINQ 233
Score = 29.3 bits (64), Expect(2) = e-100
Identities = 13/16 (81%), Positives = 13/16 (81%)
Frame = -3
Query: 332 EQLLLHPFITKYETAK 347
EQL PFITKYETAK
Sbjct: 797 EQLFCTPFITKYETAK 750
>BI308090 similar to GP|21655213|gb mitogen-activated protein kinase kinase
{Suaeda maritima subsp. salsa}, partial (23%)
Length = 482
Score = 217 bits (552), Expect(2) = 9e-72
Identities = 107/107 (100%), Positives = 107/107 (100%)
Frame = +3
Query: 38 GGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSV 97
GGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSV
Sbjct: 162 GGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSV 341
Query: 98 VQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLCEAPCYEGLVE 144
VQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLCEAPCYEGLVE
Sbjct: 342 VQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLCEAPCYEGLVE 482
Score = 72.0 bits (175), Expect(2) = 9e-72
Identities = 34/34 (100%), Positives = 34/34 (100%)
Frame = +1
Query: 1 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYT 34
MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYT
Sbjct: 58 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYT 159
>TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sativa subsp.
x varia}, complete
Length = 1677
Score = 129 bits (324), Expect(2) = 2e-59
Identities = 68/155 (43%), Positives = 91/155 (57%), Gaps = 8/155 (5%)
Frame = +3
Query: 223 KITDFGISAGLENSVAMCATFVGTVTYMSPERIRNES--YSYPADIWSLGLALLESGTGE 280
++ DFG+SA +E++ TF+GT YMSPERI Y+Y +DIWSLGL LLE G
Sbjct: 840 RLPDFGVSAIMESTSGQANTFIGTYNYMSPERINGSQRGYNYKSDIWSLGLILLECAMGR 1019
Query: 281 FPYTAN------EGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQL 334
FPYT E L+ I+D P PS E+FS EFCSF+ ACLQKDP +R +A++L
Sbjct: 1020FPYTPPDQSERWESIFELIETIVDKPPPSAPSEQFSSEFCSFISACLQKDPGSRLSAQEL 1199
Query: 335 LLHPFITKYETAKVDLPGFVRSVFDPTQRMKDLAD 369
+ PFI+ Y+ VDL + P + + D
Sbjct: 1200MELPFISMYDDLHVDLSAYFSDAGSPLATL*KMID 1304
Score = 119 bits (297), Expect(2) = 2e-59
Identities = 77/198 (38%), Positives = 117/198 (58%), Gaps = 1/198 (0%)
Frame = +2
Query: 31 DSYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAI 90
+S TF DG +LL GV ++E ++ ++ D + ++ I +
Sbjct: 302 ESGTFKDG---DLLVNRDGVRIVSETEVEAPPPIKATDN--------QLSLADIDIVKVV 448
Query: 91 GSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
G G VVQ H T++ ALK I + E+ R+Q+ E++ A C +V + +F
Sbjct: 449 GKGNGGVVQLVQHKWTNQFFALKIIQMNIEESVRKQIAKELKINQAAQC-PYVVVCYQSF 625
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
Y D+G ISI LEYMDGGS+AD+L+ +TIPEP LS++ +++L+GL YLH R+++HRD+
Sbjct: 626 Y--DNGVISIILEYMDGGSMADLLKKVKTIPEPYLSAICKQVLKGLIYLHHERHIIHRDL 799
Query: 210 KPANLLVNLKGEPKITDF 227
KP+NLL+N GE KIT F
Sbjct: 800 KPSNLLINHTGEVKIT*F 853
>BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabacum}, partial
(60%)
Length = 648
Score = 200 bits (508), Expect = 1e-51
Identities = 104/218 (47%), Positives = 145/218 (65%), Gaps = 6/218 (2%)
Frame = +2
Query: 126 LLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILS 185
++ E++ + C +V +H +FY ++G IS+ LEYMD GSL D++R TI EP L+
Sbjct: 2 IVQELKINQASQCPHVVVCYH-SFY--NNGVISLVLEYMDRGSLVDVIRQVNTILEPYLA 172
Query: 186 SMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVG 245
+ +++L+GL YLH R+++HRDIKP+NLLVN KGE KITDFG+SA L +++ TFVG
Sbjct: 173 VVCKQVLQGLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFGVSAMLASTMGQRDTFVG 352
Query: 246 TVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGP------VNLMLQILDD 299
T YMSPERI +Y Y DIWSLG+ +LE G FPY +E L+ I++
Sbjct: 353 TYNYMSPERISGSTYDYSCDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLQAIVES 532
Query: 300 PSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLH 337
P PS ++FSPEFCSFV +C++KDP R T+ +LL H
Sbjct: 533 PPPSAPPDQFSPEFCSFVSSCIKKDPRERSTSLELLDH 646
>TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase T9L24.32
[imported] - Arabidopsis thaliana, partial (71%)
Length = 1119
Score = 175 bits (443), Expect = 4e-44
Identities = 110/264 (41%), Positives = 153/264 (57%), Gaps = 11/264 (4%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEK--RQQLLTEIRTLCEAPCYEGLVEFHG 147
+G G V + H T + ALK IN ++ + R++ LTE+ L A +V++HG
Sbjct: 219 LGHGNGGTVYKVRHKLTSIIYALK-INHYDSDPTTRRRALTEVNILRRATDCTNVVKYHG 395
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHR 207
+F P +G + I +EYMD GSL L+ T E LS++ + +L GL+YLH R + HR
Sbjct: 396 SFEKP-TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHA-RNIAHR 569
Query: 208 DIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY-----SY 262
DIKP+N+LVN+K E KI DFG+S + ++ C ++VGT YMSPER E Y +
Sbjct: 570 DIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGF 749
Query: 263 PADIWSLGLALLESGTGEFPY-TANEGP--VNLMLQI-LDDPSPSPSKEKFSPEFCSFVD 318
ADIWSLGL L E G FP+ + + P +LM I DP P E S EF +FV+
Sbjct: 750 SADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLP--ETASSEFRNFVE 923
Query: 319 ACLQKDPDNRPTAEQLLLHPFITK 342
CL+K+ R +A QLL HPF+ K
Sbjct: 924 CCLKKESGERWSAAQLLTHPFLCK 995
>TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase kinsae
{Medicago sativa subsp. x varia}, complete
Length = 1546
Score = 161 bits (408), Expect = 5e-40
Identities = 106/289 (36%), Positives = 157/289 (53%), Gaps = 8/289 (2%)
Frame = +2
Query: 83 EMRIFGAIGSGASSVVQRAMHIPTHRVIALKKI-NIFEKEKRQQLLTEIRTLCEAPCYEG 141
E+ IGSG+ V + +H R ALK I E+ R+Q+ EI+ L +
Sbjct: 464 ELERLNRIGSGSGGTVYKVVHRINGRAYALKVIYGHHEESVRRQIHREIQILRDVDDVN- 640
Query: 142 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP-EPILSSMFQKLLRGLSYLHG 200
+V+ H + + +I + LEYMDGGSL + IP E L+ + +++LRGL+YLH
Sbjct: 641 VVKCHEMY--DHNAEIQVLLEYMDGGSLEG-----KHIPQENQLADVARQILRGLAYLHR 799
Query: 201 VRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY 260
R++VHRDIKP+NLL+N + + KI DFG+ L ++ C + VGT+ YMSPERI +
Sbjct: 800 -RHIVHRDIKPSNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDIN 976
Query: 261 -----SYPADIWSLGLALLESGTGEFPY-TANEGPVNLMLQILDDPSPSPSKEKFSPEFC 314
+Y DIWSLG+++LE G FP+ +G ++ + P + SPEF
Sbjct: 977 DGQYDAYAGDIWSLGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQPPEAPTTASPEFR 1156
Query: 315 SFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQR 363
FV CLQ+DP R TA +LL HPF+ + + P + + P R
Sbjct: 1157DFVSRCLQRDPSRRWTASRLLSHPFLVRNGSNHNQSPPNMHQLLPPPPR 1303
>TC83271 similar to GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago
sativa subsp. x varia}, partial (42%)
Length = 683
Score = 155 bits (391), Expect = 5e-38
Identities = 78/154 (50%), Positives = 105/154 (67%), Gaps = 10/154 (6%)
Frame = +1
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIR----NESYSYPAD 265
KP+N+L+N +GE KITDFG+SA +E++ TF+GT YMSPERI+ + Y+Y +D
Sbjct: 10 KPSNILINHRGEVKITDFGVSAIMESTSGQANTFIGTYNYMSPERIKASDSEQGYNYKSD 189
Query: 266 IWSLGLALLESGTGEFPYTA---NEGPVNLML---QILDDPSPSPSKEKFSPEFCSFVDA 319
IWSLGL LL+ TG+FPYT +EG NL L I++ PSPS ++ SPEFCSF+ A
Sbjct: 190 IWSLGLMLLKCATGKFPYTPPDNSEGWENLFLLIEAIVEKPSPSAPPDECSPEFCSFISA 369
Query: 320 CLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGF 353
CLQK+P +RP+ LL HPF+ Y+ VDL +
Sbjct: 370 CLQKNPRDRPSTRNLLRHPFVNMYDDLHVDLSDY 471
>TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 protein kinase
{Brassica napus}, partial (52%)
Length = 2266
Score = 140 bits (352), Expect = 2e-33
Identities = 92/276 (33%), Positives = 152/276 (54%), Gaps = 6/276 (2%)
Frame = +2
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIF-----EKEKRQQLLTEIRTLCEAPCYEGLVE 144
+G G V + ++ A+K++ + KE +QL EI L + + +V+
Sbjct: 989 LGRGTFGHVYLGFNSENGQMCAIKEVRVGCDDQNSKECLKQLHQEIDLLNQLS-HPNIVQ 1165
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYL 204
+ G+ +S +S+ LEY+ GGS+ +L+ + EP++ + ++++ GL+YLHG R
Sbjct: 1166 YLGSELGEES--LSVYLEYVSGGSIHKLLQEYGPFKEPVIQNYTRQIVSGLAYLHG-RNT 1336
Query: 205 VHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNES-YSYP 263
VHRDIK AN+LV+ GE K+ DFG++ + ++ +M + F G+ +M+PE + N + YS P
Sbjct: 1337 VHRDIKGANILVDPNGEIKLADFGMAKHITSAASMLS-FKGSPYWMAPEVVMNTNGYSLP 1513
Query: 264 ADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
DIWSLG L+E + P++ EG V + +I + E S + +F+ CLQ+
Sbjct: 1514 VDIWSLGCTLIEMAASKPPWSQYEG-VAAIFKIGNSKDMPIIPEHLSNDAKNFIMLCLQR 1690
Query: 324 DPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFD 359
DP RPTA++LL HPFI K +FD
Sbjct: 1691 DPSARPTAQKLLEHPFIRDQSATKAATRDVSSYMFD 1798
>TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (34%)
Length = 1847
Score = 117 bits (292), Expect = 1e-26
Identities = 86/265 (32%), Positives = 143/265 (53%), Gaps = 14/265 (5%)
Frame = +1
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKR-----QQLLTEIRTLCEAPCYEGLVE 144
IG G+ V A ++ T ALK++++ + + +QL EIR L + + +VE
Sbjct: 1042 IGRGSFGSVYHATNLETGASCALKEVDLVPDDPKSTDCIKQLDQEIRILGQLH-HPNIVE 1218
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMH-RTIPEPILSSMFQKLLRGLSYLHGVRY 203
++G+ D ++ I +EY+ GSL ++ H + E ++ + + +L GL+YLH +
Sbjct: 1219 YYGSEVVGD--RLCIYMEYVHPGSLQKFMQDHCGVMTESVVRNFTRHILSGLAYLHSTK- 1389
Query: 204 LVHRDIKPANLLVNLKGEPKITDFGISAGL-ENSVAMCATFVGTVTYMSPERIR------ 256
+HRDIK ANLLV+ G K+ DFG+S L E S + + G+ +M+PE +
Sbjct: 1390 TIHRDIKGANLLVDASGIVKLADFGVSKILTEKSYEL--SLKGSPYWMAPELMMAAMKNE 1563
Query: 257 -NESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCS 315
N + + DIWSLG ++E TG+ P++ G M ++L P + SPE
Sbjct: 1564 TNPTVAMAVDIWSLGCTIIEMLTGKPPWSEFPGH-QAMFKVLHRSPDIP--KTLSPEGQD 1734
Query: 316 FVDACLQKDPDNRPTAEQLLLHPFI 340
F++ C Q++P +RP+A LL HPF+
Sbjct: 1735 FLEQCFQRNPADRPSAAVLLTHPFV 1809
>TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated protein
kinase {Arabidopsis thaliana}, partial (26%)
Length = 789
Score = 113 bits (282), Expect = 2e-25
Identities = 69/170 (40%), Positives = 100/170 (58%), Gaps = 3/170 (1%)
Frame = +2
Query: 176 HRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLE- 234
H + + +S+ +++L GL YLH R +VHRDIK AN+LV+ G K+ DFG+S ++
Sbjct: 2 HEKLRDSQVSAYTRQILHGLKYLHD-RNIVHRDIKCANILVDANGSVKVADFGLS*AIKL 178
Query: 235 NSVAMCATFVGTVTYMSPERIRN--ESYSYPADIWSLGLALLESGTGEFPYTANEGPVNL 292
N V C GT +M+PE +R + Y PADIWSLG +LE TG+ PY E ++
Sbjct: 179 NDVKSCQ---GTAFWMAPEVVRGKVKGYGLPADIWSLGCTVLEMLTGQVPYAPME-CISA 346
Query: 293 MLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITK 342
M +I P P + S + F+ CL+ +PD+RPTA QLL H F+ +
Sbjct: 347 MFRIGKGELP-PVPDTLSRDARDFILQCLKVNPDDRPTAAQLLDHKFVQR 493
>TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein kinase
{Arabidopsis thaliana}, partial (44%)
Length = 921
Score = 111 bits (278), Expect = 6e-25
Identities = 69/210 (32%), Positives = 114/210 (53%), Gaps = 9/210 (4%)
Frame = +2
Query: 139 YEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMH--RTIPEPILSSMFQKLLRGLS 196
+ L+ H +F S + + + +M GGS I++ EP+++++ +++L+ L
Sbjct: 59 HPNLLRAHCSFTAGHS--LWVVMPFMSGGSCLHIMKSSFPEGFDEPVIATVLREVLKALV 232
Query: 197 YLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENS---VAMCATFVGTVTYMSPE 253
YLH + +HRD+K N+L++ G K+ DFG+SA + ++ TFVGT +M+PE
Sbjct: 233 YLHAHGH-IHRDVKAGNILLDANGSVKMADFGVSACMFDTGDRQRSRNTFVGTPCWMAPE 409
Query: 254 RIRN-ESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEK---F 309
++ Y + ADIWS G+ LE G P++ P+ ++L L + P E+ F
Sbjct: 410 VMQQLHGYDFKADIWSFGITALELAHGHAPFSKYP-PMKVLLMTLQNAPPGLDYERDKRF 586
Query: 310 SPEFCSFVDACLQKDPDNRPTAEQLLLHPF 339
S F V CL KDP RP++E+LL H F
Sbjct: 587 SKSFKELVATCLVKDPKKRPSSEKLLKHHF 676
>TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Brassica
napus}, partial (34%)
Length = 851
Score = 96.3 bits (238), Expect(2) = 3e-22
Identities = 58/169 (34%), Positives = 98/169 (57%), Gaps = 1/169 (0%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKR-QQLLTEIRTLCEAPCYEGLVEFHGA 148
IG G+ V + ++ +A+K I++ E E + EI L + C + E++G+
Sbjct: 207 IGQGSFGDVYKGFDKELNKEVAIKVIDLEESEDDIDDIQKEISVLSQCRC-PYITEYYGS 383
Query: 149 FYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRD 208
F + ++ I +EYM GGS+AD+L+ + E ++ + + LL + YLH +HRD
Sbjct: 384 FL--NQTKLWIIMEYMAGGSVADLLQSGPPLDEMSIAYILRDLLHAVDYLHN-EGKIHRD 554
Query: 209 IKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRN 257
IK AN+L++ G+ K+ DFG+SA L +++ TFVGT +M+PE I+N
Sbjct: 555 IKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQN 701
Score = 26.9 bits (58), Expect(2) = 3e-22
Identities = 10/23 (43%), Positives = 15/23 (64%)
Frame = +1
Query: 257 NESYSYPADIWSLGLALLESGTG 279
++ Y+ ADIWSLG+ +E G
Sbjct: 703 SKGYNEKADIWSLGITAIEMAKG 771
>TC79149 similar to PIR|T46059|T46059 MAP kinase - Arabidopsis thaliana,
partial (63%)
Length = 1671
Score = 102 bits (253), Expect = 5e-22
Identities = 74/268 (27%), Positives = 134/268 (49%), Gaps = 6/268 (2%)
Frame = +3
Query: 85 RIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFE----KEKRQQLLTEIRTLCEAPCYE 140
R +G GA V +A+ +A ++ + E + Q+L +E+ L +
Sbjct: 423 RFGDVLGKGAMKTVYKAIDEVLGIEVAWNQVRLNEVLNTPDDLQRLYSEVHLLSTLK-HR 599
Query: 141 GLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHG 200
++ F+ ++ D+ + E GSL + R ++ + + S +++L+GL YLHG
Sbjct: 600 SIMRFYTSWIDIDNKNFNFVTEMFTSGSLREYRRKYKRVSLQAIKSWARQILQGLVYLHG 779
Query: 201 -VRYLVHRDIKPANLLVN-LKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNE 258
++HRD K N+ VN G+ KI D G++A L+ S + + +GT +M+PE + E
Sbjct: 780 HDPPVIHRDPKCDNIFVNGHLGQVKIGDLGLAAILQGSQS-AHSVIGTPEFMAPE-MYEE 953
Query: 259 SYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVD 318
Y+ AD++S G+ +LE T ++PY+ P + ++ P E F+
Sbjct: 954 EYNELADVYSFGMCVLEMLTSDYPYSECTNPAQIYKKVTSGKLPMSFFRIEDGEARRFIG 1133
Query: 319 ACLQKDPDNRPTAEQLLLHPFITKYETA 346
CL+ NRP+A+ LLL PF++ +T+
Sbjct: 1134KCLE-PAANRPSAKDLLLEPFLSTDDTS 1214
>TC88048 similar to GP|20302596|dbj|BAB91125. Ser/Thr kinase {Arabidopsis
thaliana}, partial (61%)
Length = 2349
Score = 100 bits (250), Expect = 1e-21
Identities = 80/270 (29%), Positives = 134/270 (49%), Gaps = 10/270 (3%)
Frame = +1
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFE----KEKRQQLLTEIRTLCEAPCYEGLVEF 145
+G GAS V RA +A ++ +++ E ++L EI L + ++ +++F
Sbjct: 100 LGKGASKTVYRAFDEYQGIEVAWNQVKLYDFLQSPEDLERLYCEIHLL-KTLKHKNIMKF 276
Query: 146 HGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRY-L 204
+ ++ + I+ E G+L H+ + + ++LRGL YLH +
Sbjct: 277 YTSWVDTANRNINFVTEMFTSGTLRQYRLKHKRVNIRAVKHWCIQILRGLLYLHSHDPPV 456
Query: 205 VHRDIKPANLLVN-LKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYP 263
+HRD+K N+ +N +GE KI D G++A L S A A VGT +M+PE + ESY+
Sbjct: 457 IHRDLKCDNIFINGNQGEVKIGDLGLAAILRKSHA--AHCVGTPEFMAPE-VYEESYNEL 627
Query: 264 ADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
DI+S G+ +LE T E+PY+ P + +++ P + PE FVD CL
Sbjct: 628 VDIYSFGMCILEMVTFEYPYSECTHPAQIYKKVISGKKPDALYKVKDPEVRQFVDKCL-A 804
Query: 324 DPDNRPTAEQLLLHPFIT----KYETAKVD 349
R +A++LL PF+ +Y+ VD
Sbjct: 805 TVSLRLSAKELLDDPFLQIDDYEYDLRPVD 894
>TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (45%)
Length = 976
Score = 98.2 bits (243), Expect = 7e-21
Identities = 59/189 (31%), Positives = 101/189 (53%), Gaps = 2/189 (1%)
Frame = +1
Query: 157 ISIALEYMDGGSLADILRMHRTI--PEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANL 214
+ I + Y +GG +A+ ++ + PE L +LL L YLH V +++HRD+K +N+
Sbjct: 304 VCIIIGYCEGGDMAETVKKANGVNFPEEKLCKWLVQLLMALDYLH-VNHILHRDVKCSNI 480
Query: 215 LVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALL 274
+ + ++ DFG+ A L S + ++ VGT +YM PE + + Y +DIWSLG +
Sbjct: 481 FLTKNQDIRLGDFGL-AKLLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCVY 657
Query: 275 ESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQL 334
E + A + + ++ ++ SP +S F V + L+K+P+ RPTA +L
Sbjct: 658 EMAAHRPAFKAFD--IQALIHKINKSIVSPLPTMYSSAFRGLVKSMLRKNPELRPTAGEL 831
Query: 335 LLHPFITKY 343
L HP + Y
Sbjct: 832 LNHPHLQPY 858
>TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine/threonine
protein kinase {Glycine max}, partial (90%)
Length = 2194
Score = 94.0 bits (232), Expect = 1e-19
Identities = 88/317 (27%), Positives = 138/317 (42%), Gaps = 26/317 (8%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKIN---IFEKEKRQQLLTEIRTLCEAPCYEGLVEFH 146
+G G+ V+ A H+ T +A+K +N I E +++ EI+ L + ++ +
Sbjct: 132 LGIGSFGKVKIAEHVLTGHKVAIKILNRRKIKNMEMEEKVRREIKIL-RLFMHHHIIRLY 308
Query: 147 GAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
TP I + +EY+ G L D + + E S FQ+++ G+ Y H +VH
Sbjct: 309 EVVETPTD--IYVVMEYVKSGELFDYIVEKGRLQEDEARSFFQQIISGVEYCHR-NMVVH 479
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYP-AD 265
RD+KP NLL++ K KI DFG+S + T G+ Y +PE I + Y+ P D
Sbjct: 480 RDLKPENLLLDSKWSVKIADFGLS-NIMRDGHFLKTSCGSPNYAAPEVISGKLYAGPEVD 656
Query: 266 IWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDP 325
+WS G+ L G P+ +E NL +I PS SP + L DP
Sbjct: 657 VWSCGVILYALLCGTLPFD-DENIPNLFKKIKGGIYTLPS--HLSPGARDLIPRLLVVDP 827
Query: 326 DNRPTAEQLLLHPFI--------------TKYETAKVDLPGFVRSVFDPTQRMKDLADML 371
R T ++ HP+ T + K+D ++ V + + L D L
Sbjct: 828 MKRMTIPEIRQHPWFQLHLPRYLAVPPPDTLQQAKKID-EEILQEVVNRGFAREPLVDSL 1004
Query: 372 --------TIHYYLLFD 380
T+ YYLL D
Sbjct: 1005KNRVQNEGTVTYYLLLD 1055
>TC79461 similar to GP|9758254|dbj|BAB08753.1 contains similarity to
NRK-related kinase~gene_id:MWC10.1 {Arabidopsis
thaliana}, partial (88%)
Length = 1230
Score = 92.8 bits (229), Expect = 3e-19
Identities = 70/257 (27%), Positives = 129/257 (49%), Gaps = 7/257 (2%)
Frame = +1
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIF----EKEKRQQLLTEIRTLCEAPCYEGLVEF 145
+G GA V RA +A ++ + E ++L +E+R L + + ++E
Sbjct: 142 LGCGAVKKVYRAFDQEEGIEVAWNQVKLRNFCDEPAMVERLYSEVRLL-RSLTNKNIIEL 318
Query: 146 HGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVR-YL 204
+ + + ++ E G+L + + HR + L +++L+GL+YLH +
Sbjct: 319 YSVWSDDRNNTLNFITEVCTSGNLREYRKKHRHVSMKALKKWSRQILKGLNYLHTHEPCI 498
Query: 205 VHRDIKPANLLVNLK-GEPKITDFGISAGL-ENSVAMCATFVGTVTYMSPERIRNESYSY 262
+HRD+ +N+ VN G+ KI D G++A + +N +A T +GT +M+PE + +E Y+
Sbjct: 499 IHRDLNCSNVFVNGNVGQVKIGDLGLAAIVGKNHIA--HTILGTPEFMAPE-LYDEDYTE 669
Query: 263 PADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQ 322
DI+S G+ +LE T E PY+ + + ++ P+ + E F++ CL
Sbjct: 670 LVDIYSFGMCVLEMVTLEIPYSECDNVAKIYKKVSSGIRPAAMNKVKDSEVKEFIERCLA 849
Query: 323 KDPDNRPTAEQLLLHPF 339
+ P RP+A +LL PF
Sbjct: 850 Q-PRARPSAAELLKDPF 897
>TC91968 similar to PIR|T02550|T02550 NPK1-related protein kinase homolog
T26B15.7 - Arabidopsis thaliana, partial (20%)
Length = 662
Score = 92.0 bits (227), Expect = 5e-19
Identities = 66/213 (30%), Positives = 114/213 (52%), Gaps = 6/213 (2%)
Frame = +2
Query: 90 IGSGASSVVQRAMHIPTH-RVIALKKINIFEKE---KRQQLLTEIRTLCEAPCYEGLVEF 145
IG G+++ V A + V A+K + E K QQ+LT+++ C + +V +
Sbjct: 17 IGRGSTATVYVADRCHSGGEVFAVKSTELHRSELLKKEQQILTKLK------CPQ-IVSY 175
Query: 146 HGAFYTPDSGQ--ISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRY 203
G T ++G ++ +EY G+L+D +R + E ++ +++L GL+YLH +
Sbjct: 176 QGCEVTFENGVQLFNLFMEYAPKGTLSDAVRNGNGMEEAMVGFYARQILLGLNYLH-LNG 352
Query: 204 LVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYP 263
+VH D+K N+LV +G KI+DFG + +E + + GT +M+PE R E Y
Sbjct: 353 IVHCDLKGQNVLVTEQGA-KISDFGCARRVEEELVIS----GTPAFMAPEVARGEEQGYA 517
Query: 264 ADIWSLGLALLESGTGEFPYTANEGPVNLMLQI 296
+D+W+LG LLE TG+ P+ P ++ +I
Sbjct: 518 SDVWALGCTLLEMITGKMPWQGFSDPAAVLYRI 616
>TC84173 similar to PIR|A71407|A71407 probable Ste20-like kinase -
Arabidopsis thaliana, partial (41%)
Length = 706
Score = 91.3 bits (225), Expect = 8e-19
Identities = 71/218 (32%), Positives = 114/218 (51%), Gaps = 24/218 (11%)
Frame = +2
Query: 77 YRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRV-IALKKINIFE-KEKRQQLLTEIRTLC 134
Y S +I IG+G S+VV +A+ IP + +A+K I++ + + E +TL
Sbjct: 65 YPLDSSSYKIVDEIGAGNSAVVYKAICIPINSTPVAIKSIDLDRSRPDLDDVRREAKTL- 241
Query: 135 EAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILR--MHRTIPEPILSSMFQKLL 192
+ +++ H +F T D+ ++ + + +M GGSL I+ + E ++ + + L
Sbjct: 242 SLLSHPNILKAHCSF-TVDN-RLWVVMPFMAGGSLQSIISHSFQNGLTEQSIAVILKDTL 415
Query: 193 RGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGL---ENSVAMCAT------- 242
LSYLHG +L HRDIK N+LV+ G K+ DFG+SA + NSV C++
Sbjct: 416 NALSYLHGQGHL-HRDIKSGNILVDSNGLVKLADFGVSASIYESNNSVGACSSYSSSSSN 592
Query: 243 ---------FVGTVTYMSPERIRNES-YSYPADIWSLG 270
F GT +M+PE I + + YS+ ADIWS G
Sbjct: 593 SSSSHIFTDFAGTPYWMAPEGIHSHNGYSFKADIWSFG 706
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,233,292
Number of Sequences: 36976
Number of extensions: 239511
Number of successful extensions: 1849
Number of sequences better than 10.0: 505
Number of HSP's better than 10.0 without gapping: 1563
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1611
length of query: 519
length of database: 9,014,727
effective HSP length: 100
effective length of query: 419
effective length of database: 5,317,127
effective search space: 2227876213
effective search space used: 2227876213
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146560.10


