

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.11 - phase: 0
(367 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
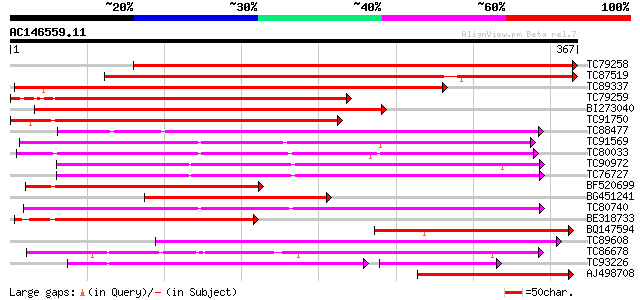
Score E
Sequences producing significant alignments: (bits) Value
TC79258 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 571 e-163
TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDS... 392 e-109
TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 372 e-103
TC79259 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif... 321 3e-88
BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/ac... 320 7e-88
TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif... 291 2e-79
TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase p... 220 6e-58
TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protei... 218 2e-57
TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 210 6e-55
TC90972 similar to GP|10638955|emb|CAB81548. putative proline-ri... 202 1e-52
TC76727 similar to GP|10638955|emb|CAB81548. putative proline-ri... 202 2e-52
BF520699 similar to PIR|T04521|T04 proline-rich protein APG homo... 197 5e-51
BG451241 similar to GP|6721157|gb| putative GDSL-motif lipase/ac... 194 5e-50
TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif lipase/h... 194 6e-50
BE318733 similar to GP|21592967|gb putative GDSL-motif lipase/ac... 182 2e-46
BQ147594 weakly similar to GP|6721157|gb| putative GDSL-motif li... 179 1e-45
TC89608 similar to PIR|T01143|T01143 probable GDSL-motif lipase/... 171 3e-43
TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20... 160 1e-39
TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein... 135 6e-38
AJ498708 similar to GP|21592967|gb putative GDSL-motif lipase/ac... 150 8e-37
>TC79258 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(73%)
Length = 1067
Score = 571 bits (1471), Expect = e-163
Identities = 284/287 (98%), Positives = 285/287 (98%)
Frame = +3
Query: 81 LTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQ 140
+ ERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQ
Sbjct: 24 IAGERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQ 203
Query: 141 YQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISE 200
YQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISE
Sbjct: 204 YQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISE 383
Query: 201 YKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMIT 260
YKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMIT
Sbjct: 384 YKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMIT 563
Query: 261 QLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPN 320
QLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPN
Sbjct: 564 QLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPN 743
Query: 321 RNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDSMV 367
RNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDSMV
Sbjct: 744 RNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDSMV 884
>TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(76%)
Length = 1084
Score = 392 bits (1007), Expect = e-109
Identities = 192/308 (62%), Positives = 242/308 (78%), Gaps = 2/308 (0%)
Frame = +2
Query: 62 DFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDT 121
DFPT +PTGRFSNGLN+PDL S+ LG P LPYLSP L G ++L GANFASAG+GILNDT
Sbjct: 2 DFPTRQPTGRFSNGLNVPDLISKELGSSPPLPYLSPKLRGHRMLNGANFASAGIGILNDT 181
Query: 122 GFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPF 181
GFQF+++I + KQLD F +YQ+++S IG + AK+L+N A++LI GGNDFVNNYYLVP
Sbjct: 182 GFQFIEVIRMYKQLDFFEEYQKRVSDLIGKKEAKKLINGALILITCGGNDFVNNYYLVPN 361
Query: 182 SARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCD 241
S RSRQ++LP YVTYL+SEYKKIL+RLY LGARRVLV+GTGPMGCAPA LA+ +G+C
Sbjct: 362 SLRSRQYALPEYVTYLLSEYKKILRRLYHLGARRVLVSGTGPMGCAPAALAIGGTDGECA 541
Query: 242 AELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFG--FVTAKDAC 299
EL AASLYNP+LVQ+IT+LN++IG DVF +N + + FG F T+K AC
Sbjct: 542 PELQLAASLYNPKLVQLITELNQQIGSDVFSVLNIDALSL--------FGNEFKTSKVAC 697
Query: 300 CGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLST 359
CGQG +NGIGLCT S +C NR+ + FWDAFHPSE+A+++IV+Q+ GS + PMNLST
Sbjct: 698 CGQGPYNGIGLCTLASSICQNRDDHLFWDAFHPSERANKMIVKQIMTGSTDVIYPMNLST 877
Query: 360 VLAMDSMV 367
+LA+DS +
Sbjct: 878 ILALDSKI 901
>TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(65%)
Length = 881
Score = 372 bits (954), Expect = e-103
Identities = 184/283 (65%), Positives = 222/283 (78%), Gaps = 3/283 (1%)
Frame = +1
Query: 4 SLMLCCSYILMINLFVG--FDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGI 61
S+ S++ ++ L VG F R F VFGDS+ DNGNNN+L TTARADAPPYGI
Sbjct: 31 SMASLSSFVALVILVVGGIFVHEIEAIPRTFLVFGDSLVDNGNNNYLATTARADAPPYGI 210
Query: 62 DF-PTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILND 120
D+ P+H PTGRFSNG NIPD+ S++LG EP+LPYLSP L GEKLLVGANFASAG+GILND
Sbjct: 211 DYQPSHRPTGRFSNGYNIPDIISQKLGAEPTLPYLSPELRGEKLLVGANFASAGIGILND 390
Query: 121 TGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVP 180
TG QF+ II + +Q + F +YQ +LSA IGA AK VN+A+VLI +GGNDFVNNYYLVP
Sbjct: 391 TGIQFINIIRMYRQYEYFQEYQSRLSALIGASQAKSRVNQALVLITVGGNDFVNNYYLVP 570
Query: 181 FSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDC 240
+SARSRQ+ LP YV YLISEY+K+LQ+LYDLGARRVLVTGTGPMGC P+E+A + RNG C
Sbjct: 571 YSARSRQYPLPEYVKYLISEYQKLLQKLYDLGARRVLVTGTGPMGCVPSEIAQRGRNGQC 750
Query: 241 DAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDF 283
EL RA+SL+NPQL M+ LN++IG DVFIA N K H++F
Sbjct: 751 STELQRASSLFNPQLENMLLGLNKKIGRDVFIAANTGKTHLNF 879
>TC79259 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(52%)
Length = 712
Score = 321 bits (822), Expect = 3e-88
Identities = 166/221 (75%), Positives = 193/221 (87%)
Frame = +3
Query: 1 MASSLMLCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYG 60
MA+SL++ ++ +MI+ FVG AYAQP RAF VFGDS+ D+GNN+FL TTARAD PYG
Sbjct: 66 MATSLVI--AFCVMIS-FVG--CAYAQP-RAFGVFGDSLVDSGNNDFLATTARADNYPYG 227
Query: 61 IDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILND 120
ID+P+H PTGRFSNG NIPDL S LGLEP+LPYLSPLLVGEKLL+GANFASAG+GILND
Sbjct: 228 IDYPSHRPTGRFSNGYNIPDLISLELGLEPTLPYLSPLLVGEKLLIGANFASAGIGILND 407
Query: 121 TGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVP 180
TGFQF+ II I KQL LF YQ+++SA IG+EGA+ LVN+A+VLI LGGNDFVNNYYLVP
Sbjct: 408 TGFQFIHIIRIYKQLRLFELYQKRVSAHIGSEGARNLVNRALVLITLGGNDFVNNYYLVP 587
Query: 181 FSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGT 221
FSARSRQFSLP+YV YLISEY+K+L+RLYDLGARRVLVTGT
Sbjct: 588 FSARSRQFSLPDYVRYLISEYRKVLRRLYDLGARRVLVTGT 710
>BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (60%)
Length = 692
Score = 320 bits (819), Expect = 7e-88
Identities = 158/229 (68%), Positives = 184/229 (79%), Gaps = 1/229 (0%)
Frame = +2
Query: 17 LFVGFDLAYAQPK-RAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNG 75
L V F+ Q + RAFFVFGDS+ DNGNNN+L TTARAD+ PYGID+PTH TGRFSNG
Sbjct: 2 LLVNFNTVVPQVEARAFFVFGDSLVDNGNNNYLATTARADSYPYGIDYPTHRATGRFSNG 181
Query: 76 LNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQL 135
LN+PDL SER+G +P+LPYLSP L GE LLVGANFASAG+GILNDTG QF II I +QL
Sbjct: 182 LNMPDLISERIGSQPTLPYLSPELNGEALLVGANFASAGIGILNDTGIQFFNIIRITRQL 361
Query: 136 DLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVT 195
F QYQQ++SA IG E +LVN+A+ L+ LGGNDFVNNY+LVPFSARSRQF LP+YV
Sbjct: 362 QYFEQYQQRVSALIGEEETVRLVNEALYLMTLGGNDFVNNYFLVPFSARSRQFXLPDYVV 541
Query: 196 YLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAEL 244
YLISEY+KIL RLY+LGARRVLVTGT P+GC P L +NG+C A+L
Sbjct: 542 YLISEYRKILARLYELGARRVLVTGTXPLGCVPXXLXXHXKNGECYAKL 688
>TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(50%)
Length = 686
Score = 291 bits (746), Expect = 2e-79
Identities = 151/220 (68%), Positives = 174/220 (78%), Gaps = 5/220 (2%)
Frame = +3
Query: 1 MASSLMLCCSYI-----LMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARAD 55
MA++++L YI LM+ L F AQ RAFFVFGDS+ DNGNNN+L TTARAD
Sbjct: 15 MAATMVLQSYYINVVIILMVALTSCFKGTVAQ--RAFFVFGDSLVDNGNNNYLATTARAD 188
Query: 56 APPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGV 115
APPYGID+PT PTGRFSNG NIPD S+ LG EP+LPYLSP L GE LLVGANFASAG+
Sbjct: 189 APPYGIDYPTRRPTGRFSNGYNIPDFISQALGAEPTLPYLSPELNGEALLVGANFASAGI 368
Query: 116 GILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNN 175
GILNDTG QF+ II I +QL+ F QYQQ++S IG E + LVN A+VLI LGGNDFVNN
Sbjct: 369 GILNDTGIQFINIIRIFRQLEYFQQYQQRVSGLIGPEQTQSLVNGALVLITLGGNDFVNN 548
Query: 176 YYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARR 215
YYLVPFSARSRQ++LP+YV Y+ISEYKKIL+RLY LGARR
Sbjct: 549 YYLVPFSARSRQYNLPDYVRYIISEYKKILRRLYHLGARR 668
>TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase putative
{Arabidopsis thaliana}, partial (92%)
Length = 1415
Score = 220 bits (561), Expect = 6e-58
Identities = 123/317 (38%), Positives = 179/317 (55%), Gaps = 3/317 (0%)
Frame = +2
Query: 32 FFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPS 91
+F+FGDS+ DNGNNN L + ARAD PYGIDF PTGRFSNG D +E LG +
Sbjct: 185 YFIFGDSLVDNGNNNGLQSLARADYLPYGIDFGG--PTGRFSNGKTTVDAIAELLGFDDY 358
Query: 92 LP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIG 150
+P Y S + +L G N+ASA GI +TG Q + Q+ + ++ +G
Sbjct: 359 IPPYASAS--DDAILKGVNYASAAAGIREETGRQLGARLSFSAQVQNYQSTVSQVVNILG 532
Query: 151 AEG-AKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLY 209
E A ++K I I LG ND++NNY++ F Q++ Y LI Y + L+ LY
Sbjct: 533 TEDQAASHLSKCIYSIGLGSNDYLNNYFMPQFYNTHDQYTPDEYADDLIQSYTEQLRTLY 712
Query: 210 DLGARRVLVTGTGPMGCAPAELALKSRNG-DCDAELMRAASLYNPQLVQMITQLNREIGD 268
+ AR++++ G G +GC+P ELA +S +G C E+ A ++N +L ++ Q N ++ D
Sbjct: 713 NN*ARKMVLFGIGQIGCSPNELATRSADGVTCVEEINSANQIFNNKLKGLVDQFNNQLPD 892
Query: 269 DVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWD 328
I VN++ + D I+NP A+GF CCG GR NG C P+ C NR Y FWD
Sbjct: 893 SKVIYVNSYGIFQDIISNPSAYGFSVTNAGCCGVGRNNGQFTCLPLQTPCENRREYLFWD 1072
Query: 329 AFHPSEKASRIIVQQMF 345
AFHP+E + ++ Q+ +
Sbjct: 1073AFHPTEAGNVVVAQRAY 1123
>TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protein
{Arabidopsis thaliana}, partial (60%)
Length = 1398
Score = 218 bits (556), Expect = 2e-57
Identities = 121/338 (35%), Positives = 187/338 (54%), Gaps = 4/338 (1%)
Frame = +1
Query: 7 LCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTH 66
+C ++++I + A VFGDS D+GNNN ++T +++ PYG D
Sbjct: 61 ICIVWLILITQMIMVTCNNENYVPAVIVFGDSSVDSGNNNMISTFLKSNFRPYGRDIDGG 240
Query: 67 EPTGRFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQF 125
PTGRFSNG PD SE G++ +P YL P + + G FASAG G N T
Sbjct: 241 RPTGRFSNGRIPPDFISEAFGIKSLIPAYLDPAYTIDDFVTGVCFASAGTGYDNATS-AI 417
Query: 126 LQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARS 185
L +I + K+++ + +YQ KL A IG E + +++++A+ +I LG NDF+ NYY F+
Sbjct: 418 LNVIPLWKEVEFYKEYQDKLKAHIGEEKSIEIISEALYIISLGTNDFLGNYY--GFTTLR 591
Query: 186 RQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNG--DCDAE 243
++++ Y YLI + +++LY LGAR++ +TG PMGC P E A+ G C +
Sbjct: 592 FRYTISQYQDYLIGIAENFIRQLYSLGARKLAITGLIPMGCLPLERAINIFGGFHRCYEK 771
Query: 244 LMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQG 303
A +N +L MI++LN+E+ ++ N + + D IT P +G + ACC G
Sbjct: 772 YNIVALEFNVKLENMISKLNKELPQLKALSANVYDLFNDIITRPSFYGIEEVEKACCSTG 951
Query: 304 RFNGIGLCTPISKL-CPNRNLYAFWDAFHPSEKASRII 340
LC ++ + C + + Y FWDAFHP+EK +RII
Sbjct: 952 TIEMSYLCNKMNLMTCKDASKYMFWDAFHPTEKTNRII 1065
>TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (89%)
Length = 1164
Score = 210 bits (535), Expect = 6e-55
Identities = 125/342 (36%), Positives = 187/342 (54%), Gaps = 4/342 (1%)
Frame = +1
Query: 5 LMLCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFP 64
L LC +IL + LF ++ P A VFGDS D GNNNF+ T AR++ PYG DF
Sbjct: 61 LALCSLHILCLLLFHLNKVSAKVP--AIIVFGDSSVDAGNNNFIPTVARSNFQPYGRDFQ 234
Query: 65 THEPTGRFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVGILNDTGF 123
+ TGRFSNG D +E G++ S+P YL P G +FASA G N T
Sbjct: 235 GGKATGRFSNGRIPTDFIAESFGIKESVPAYLDPKYNISDFATGVSFASAATGYDNATS- 411
Query: 124 QFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSA 183
L +I + KQL+ + YQ+ LS+ +G AK+ +++++ L+ +G NDF+ NYY +P
Sbjct: 412 DVLSVIPLWKQLEYYKDYQKNLSSYLGEAKAKETISESVHLMSMGTNDFLENYYTMP--G 585
Query: 184 RSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELA--LKSRNGDCD 241
R+ Q++ Y T+L + ++ LY LGAR++ + G PMGC P E +NG C
Sbjct: 586 RASQYTPQQYQTFLAGIAENFIRNLYALGARKISLGGLPPMGCLPLERTTNFMGQNG-CV 762
Query: 242 AELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCG 301
A A +N +L + T+LN+E+ D + N + + + I P +GF +A ACC
Sbjct: 763 ANFNNIALEFNDKLKNITTKLNQELPDMKLVFSNPYYIMLHIIKKPDLYGFESASVACCA 942
Query: 302 QGRFNGIGLCTPISKL-CPNRNLYAFWDAFHPSEKASRIIVQ 342
G F C+ S C + + + FWD+FHP+EK + I+ +
Sbjct: 943 TGMFEMGYACSRGSMFSCTDASKFVFWDSFHPTEKTNNIVAK 1068
>TC90972 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (88%)
Length = 1258
Score = 202 bits (515), Expect = 1e-52
Identities = 113/320 (35%), Positives = 167/320 (51%), Gaps = 4/320 (1%)
Frame = +3
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A FGDS D GNN++L T +A+ PPYG DF H+PTGRF NG D+T+E LG +
Sbjct: 108 AIVTFGDSAVDVGNNDYLFTLFKANYPPYGRDFVXHKPTGRFCNGKLATDITAETLGFKS 287
Query: 91 SLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQI 149
P YLSP G+ LL+GANFASA G ++ I + +QL + +YQ KLS
Sbjct: 288 YAPAYLSPQATGKNLLIGANFASAASG-YDEKAAILNHAIPLSQQLKYYKEYQSKLSKIA 464
Query: 150 GAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLY 209
G++ A ++ A+ L+ G +DF+ NYY+ P ++ + Y YL+ Y ++ LY
Sbjct: 465 GSKKAASIIKGALYLLSGGSSDFIQNYYVNPLI--NKVVTPDQYSAYLVDTYSSFVKDLY 638
Query: 210 DLGARRVLVTGTGPMGCAPAELALKS-RNGDCDAELMRAASLYNPQLVQMITQLNREIGD 268
LGAR++ VT P+GC PA L C + A +N ++ +L +++
Sbjct: 639 KLGARKIGVTSLPPLGCLPATRTLFGFHEKGCVTRINNDAQGFNKKINSATVKLQKQLPG 818
Query: 269 DVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKL--CPNRNLYAF 326
+ N +K + + +P FGF A+ CCG G L L C N Y F
Sbjct: 819 LKIVVFNIYKPLYELVQSPSKFGFAEARKGCCGTGIVETTSLLCNQKSLGTCSNATQYVF 998
Query: 327 WDAFHPSEKASRIIVQQMFI 346
WD+ HPSE A++I+ + +
Sbjct: 999 WDSVHPSEAANQILADALIV 1058
>TC76727 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (92%)
Length = 1919
Score = 202 bits (514), Expect = 2e-52
Identities = 113/320 (35%), Positives = 169/320 (52%), Gaps = 4/320 (1%)
Frame = -2
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A FGDS D GNN++L T +A+ PPYG DF +PTGRF NG D T+E LG
Sbjct: 1087 AIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTS 908
Query: 91 SLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQI 149
P YLSP G+ LL+GANFASA G ++ I + +QL+ F +YQ KL+
Sbjct: 907 FAPAYLSPQASGKNLLLGANFASAASG-YDEKAATLNHAIPLSQQLEYFKEYQGKLAQVA 731
Query: 150 GAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLY 209
G++ A ++ ++ ++ G +DFV NYY P+ ++ ++ Y +YL+ + ++ +Y
Sbjct: 730 GSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWI--NQAITVDQYSSYLLDSFTNFIKGVY 557
Query: 210 DLGARRVLVTGTGPMGCAPAELALKS-RNGDCDAELMRAASLYNPQLVQMITQLNREIGD 268
LGAR++ VT P+GC PA L C A + A +N ++ + L +++
Sbjct: 556 GLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQLPG 377
Query: 269 DVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIG-LCTPIS-KLCPNRNLYAF 326
+ + +K D + NP FGF A CCG G LC P S C N Y F
Sbjct: 376 LKIVIFDIYKPLYDLVQNPSNFGFAEAGKGCCGTGLVETTSLLCNPKSLGTCSNATQYVF 197
Query: 327 WDAFHPSEKASRIIVQQMFI 346
WD+ HPSE A++++ + I
Sbjct: 196 WDSVHPSEAANQVLADNLII 137
>BF520699 similar to PIR|T04521|T04 proline-rich protein APG homolog
F16A16.110 - Arabidopsis thaliana, partial (32%)
Length = 536
Score = 197 bits (501), Expect = 5e-51
Identities = 95/155 (61%), Positives = 125/155 (80%), Gaps = 1/155 (0%)
Frame = +1
Query: 11 YILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTG 70
++ +I+L V + A RAFFVFGDS+ D+GNNN+L TTARAD+PPYGID+P H PTG
Sbjct: 76 FVTIISLVVTMRVQAAS--RAFFVFGDSLVDSGNNNYLLTTARADSPPYGIDYPGHRPTG 249
Query: 71 RFSNGLNIPDLTSERLGL-EPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQII 129
RFSNG+N+PDL + +GL EP+LPYLSP L G++LLVGANFASAG+GILNDTG QF+ I+
Sbjct: 250 RFSNGVNLPDLIGQHIGLSEPTLPYLSPQLTGQRLLVGANFASAGIGILNDTGIQFVGIL 429
Query: 130 HIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVL 164
+ +Q LF QY+Q+LS ++GAE A ++VN+A+VL
Sbjct: 430 RMFQQFSLFEQYKQRLSGEVGAEEANRIVNEALVL 534
>BG451241 similar to GP|6721157|gb| putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (32%)
Length = 533
Score = 194 bits (493), Expect = 5e-50
Identities = 95/121 (78%), Positives = 110/121 (90%)
Frame = +3
Query: 88 LEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSA 147
LEP+LPYLSPLLVGEKLL+GANFASAG+GILNDTGFQF+ II I KQL LF YQ+++SA
Sbjct: 3 LEPTLPYLSPLLVGEKLLIGANFASAGIGILNDTGFQFIHIIRIYKQLRLFELYQKRVSA 182
Query: 148 QIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQR 207
IG+EGA+ LVN+A+VLI LGGNDFVNNYYLVPFSARSRQFSLP+YV YLISEY+K+L+
Sbjct: 183 HIGSEGARNLVNRALVLITLGGNDFVNNYYLVPFSARSRQFSLPDYVRYLISEYRKVLRV 362
Query: 208 L 208
L
Sbjct: 363 L 365
>TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif
lipase/hydrolase-like protein {Arabidopsis thaliana},
partial (94%)
Length = 1167
Score = 194 bits (492), Expect = 6e-50
Identities = 120/341 (35%), Positives = 172/341 (50%), Gaps = 4/341 (1%)
Frame = +3
Query: 10 SYILMINLFVGFDLAYAQPK-RAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEP 68
+++ + LFV F++ QP A F+FGDSV D NNN L T +++ PPYG DF P
Sbjct: 123 NFLTFLLLFVVFNVVKGQPLVPALFIFGDSVVDARNNNNLYTIVKSNFPPYGRDFNNQMP 302
Query: 69 TGRFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQ 127
TGRF NG D T+E LG P YL+ + LL GANFAS G + T +
Sbjct: 303 TGRFCNGKLAADFTAENLGFTTYPPAYLNLQEKRKNLLNGANFASGASGYFDPTA-KLYH 479
Query: 128 IIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQ 187
I + +QL+ + + Q L G A +++ AI L+ G +DFV NYY+ P +
Sbjct: 480 AISLEQQLEHYKECQNILVGVAGKSNASSIISGAIYLVRAGSSDFVQNYYINPL--LYKV 653
Query: 188 FSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELAL-KSRNGDCDAELMR 246
F+ + L+ Y +Q LY LGAR++ VT P+GC PA + L S + +C L
Sbjct: 654 FTADQFSDILMQHYTIFIQNLYALGARKIGVTTLPPLGCLPAAITLFGSHSNECVDRLNN 833
Query: 247 AASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFN 306
A +N +L L +E+ + ++ ++ D +T P GF A+ ACCG G
Sbjct: 834 DALNFNTKLNTTSQNLQKELSNLTLAVLDIYQPLHDLVTKPTENGFYEARKACCGTGLIE 1013
Query: 307 GIGLCTPIS-KLCPNRNLYAFWDAFHPSEKASRIIVQQMFI 346
LC S C N Y FWD FH SE A+ ++ + I
Sbjct: 1014TSILCNKDSIGTCANATEYVFWDGFHTSEAANNVLADDLLI 1136
>BE318733 similar to GP|21592967|gb putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (32%)
Length = 533
Score = 182 bits (462), Expect = 2e-46
Identities = 99/159 (62%), Positives = 117/159 (73%), Gaps = 1/159 (0%)
Frame = +3
Query: 4 SLMLCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDF 63
S+ LC + I L +G A RAFFVFGDS+ DNGNNNFL TTARAD+ PYGID
Sbjct: 78 SIFLC----VFITLSIGVQQIEA---RAFFVFGDSLVDNGNNNFLATTARADSYPYGIDS 236
Query: 64 PTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGF 123
TH +GRFSNGLN+PDL SE++G EP+LPYLSP L GE+LL+GANFASAG+GILNDTG
Sbjct: 237 QTHRASGRFSNGLNMPDLISEKIGSEPTLPYLSPELDGERLLIGANFASAGIGILNDTGV 416
Query: 124 QFLQIIHIGKQLDLFNQYQQK-LSAQIGAEGAKQLVNKA 161
QF+ II I QL F QYQQ+ A IG A++LVN+A
Sbjct: 417 QFINIIRITAQLAYFKQYQQEG*VALIGESEAQKLVNQA 533
>BQ147594 weakly similar to GP|6721157|gb| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(33%)
Length = 572
Score = 179 bits (455), Expect = 1e-45
Identities = 81/131 (61%), Positives = 103/131 (77%), Gaps = 2/131 (1%)
Frame = +3
Query: 237 NGDCDAELMRAASLYNPQLVQMITQLNREIG--DDVFIAVNAHKMHMDFITNPKAFGFVT 294
NG+C L A +L+NPQLV++I QLN EIG + VFI NA MH+DF++NP+A+GF T
Sbjct: 3 NGECGVPLQTATNLFNPQLVELINQLNSEIGGPNHVFIYANAFAMHLDFVSNPQAYGFET 182
Query: 295 AKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNP 354
+K ACCGQG FNG+GLCTP S LCPNR+LYAFWD FHPSE+A+R+IV + G+ YM+P
Sbjct: 183 SKQACCGQGPFNGVGLCTPASNLCPNRDLYAFWDPFHPSERANRLIVDKFMTGTTDYMHP 362
Query: 355 MNLSTVLAMDS 365
NLST++AMDS
Sbjct: 363 YNLSTLIAMDS 395
>TC89608 similar to PIR|T01143|T01143 probable GDSL-motif lipase/hydrolase
[imported] - Arabidopsis thaliana, partial (70%)
Length = 959
Score = 171 bits (434), Expect = 3e-43
Identities = 95/267 (35%), Positives = 150/267 (55%), Gaps = 4/267 (1%)
Frame = +2
Query: 95 LSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGA 154
L+P G+ +L G N+AS G GILN TG F+ I + Q+D F ++++ +G A
Sbjct: 8 LAPNATGKSILYGVNYASGGGGILNATGRIFVNRIGMDIQIDYFTITRKQIDKLLGQSKA 187
Query: 155 KQLV-NKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPN-YVTYLISEYKKILQRLYDLG 212
+ + K+I I +G NDF+NNY L S +R P+ +V +I+ ++ L RLY +
Sbjct: 188 RDFIMKKSIFSITVGANDFLNNYLLPVLSVGARISQSPDAFVDDMINHFRGQLTRLYKMD 367
Query: 213 ARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASL-YNPQLVQMITQLNREIGDDVF 271
AR+ ++ GP+GC P + + N D +L ++ YN +L M+ +LN + F
Sbjct: 368 ARKFVIGNVGPIGCIPYQKTINQLNEDECVDLANKLAIQYNGRLKDMLAELNDNLPGATF 547
Query: 272 IAVNAHKMHMDFITNPKAFGFVTAKDACCGQG-RFNGIGLCTPISKLCPNRNLYAFWDAF 330
+ N + + M+ I N +GF T+ ACCG G +F GI C P S +C +R + FWD +
Sbjct: 548 VLANVYDLVMELIKNYDKYGFTTSSRACCGNGGQFAGIIPCGPTSCICNDRYKHVFWDPY 727
Query: 331 HPSEKASRIIVQQMFIGSNLYMNPMNL 357
HPSE A+ II +Q+ G Y++P+NL
Sbjct: 728 HPSEAANIIIAKQLLDGDKRYISPVNL 808
>TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1341
Score = 160 bits (404), Expect = 1e-39
Identities = 107/345 (31%), Positives = 171/345 (49%), Gaps = 11/345 (3%)
Frame = +2
Query: 12 ILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTA--RADAPPYGIDFPTHEPT 69
I +I+ G Y + +A F+FGDS D GNNN++ TT +A+ PYG + + PT
Sbjct: 125 IAIISQTFGSKTDYYRSNKALFIFGDSFLDAGNNNYINTTTFDQANFLPYGETY-FNFPT 301
Query: 70 GRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQII 129
GRFS+G I D +E + + P+L P K G NFAS G G L +T FQ +I
Sbjct: 302 GRFSDGRLISDFIAEYVNIPLVPPFLQP--DNNKYYNGVNFASGGAGALVET-FQG-SVI 469
Query: 130 HIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARS---R 186
Q F + L ++G+ +K L++ A+ + +G ND YL PF S +
Sbjct: 470 PFKTQAINFKKVTTWLRHKLGSSDSKTLLSNAVYMFSIGSND-----YLSPFLTNSDVLK 634
Query: 187 QFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRN-GDCDAELM 245
+S YV +I + ++ ++ GA++ ++ P+GC P ++S+ G C EL
Sbjct: 635 HYSHTEYVAMVIGNFTSTIKEIHKRGAKKFVILNLPPLGCLPGTRIIQSQGKGSCLEELS 814
Query: 246 RAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRF 305
AS++N L +++ +L +++ F + + I +P +GF K ACCG G F
Sbjct: 815 SLASIHNQALYEVLLELQKQLRGFKFSLYDFNSDLSHMINHPLKYGFKEGKSACCGSGPF 994
Query: 306 NGIGLC-----TPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMF 345
G C +LC N FWD++H +E A + + QM+
Sbjct: 995 RGEYSCGGKRGEKHFELCDKPNESVFWDSYHLTESAYKQLAAQMW 1129
>TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein F3I17.10
[imported] - Arabidopsis thaliana, partial (60%)
Length = 847
Score = 135 bits (341), Expect(2) = 6e-38
Identities = 69/195 (35%), Positives = 110/195 (56%)
Frame = +2
Query: 38 SVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSP 97
S+ + GNNNFL+T A+++ PYGID+ PTGRFSNG ++ D + LG+ P+L P
Sbjct: 2 SLVEVGNNNFLSTFAKSNFYPYGIDY-NGRPTGRFSNGKSLIDFIGDMLGVPSPPPFLDP 178
Query: 98 LLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQL 157
KLL G N+AS GIL+D+G + + +QL F + + + Q
Sbjct: 179 TSTENKLLNGVNYASGSGGILDDSGRHYGDRHSMSRQLQNFERTLNQYKKMMNETALSQF 358
Query: 158 VNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVL 217
+ K+IV+++ G ND++NNY + SR +S+P + L++ + + + LY L R+
Sbjct: 359 LAKSIVIVVTGSNDYINNYLRPEYYGTSRNYSVPQFGNLLLNTFGRQILALYSLRLRKFF 538
Query: 218 VTGTGPMGCAPAELA 232
+ G GP+GC P + A
Sbjct: 539 LAGVGPLGCIPNQRA 583
Score = 39.7 bits (91), Expect(2) = 6e-38
Identities = 23/79 (29%), Positives = 33/79 (41%)
Frame = +3
Query: 240 CDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDAC 299
C + + YN L M+ Q NR+ D F+ N + + D + NP A+ F AC
Sbjct: 609 CVDSVNQMVGTYNGGLRSMVEQFNRDHSDAKFVYGNTYGVFGDILNNPAAYAFSVIDRAC 788
Query: 300 CGQGRFNGIGLCTPISKLC 318
GR C P+ C
Sbjct: 789 LCLGRK*SQISCLPMQFPC 845
>AJ498708 similar to GP|21592967|gb putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (27%)
Length = 568
Score = 150 bits (379), Expect = 8e-37
Identities = 66/101 (65%), Positives = 84/101 (82%)
Frame = +1
Query: 265 EIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLY 324
EIG +VF+ N +M +DF+ NP+A+GFVT++ ACCGQG +NG+GLCTP+S LCPNR+ Y
Sbjct: 13 EIGSNVFMGANTRQMALDFVNNPQAYGFVTSQIACCGQGPYNGLGLCTPLSNLCPNRDEY 192
Query: 325 AFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDS 365
AFWDAFHPSEKA+ +IVQQ+ G+ YM PMNLSTVLA+DS
Sbjct: 193 AFWDAFHPSEKANSLIVQQILSGTTDYMYPMNLSTVLALDS 315
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,303,486
Number of Sequences: 36976
Number of extensions: 137977
Number of successful extensions: 850
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 746
length of query: 367
length of database: 9,014,727
effective HSP length: 97
effective length of query: 270
effective length of database: 5,428,055
effective search space: 1465574850
effective search space used: 1465574850
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146559.11


