

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146557.5 - phase: 0
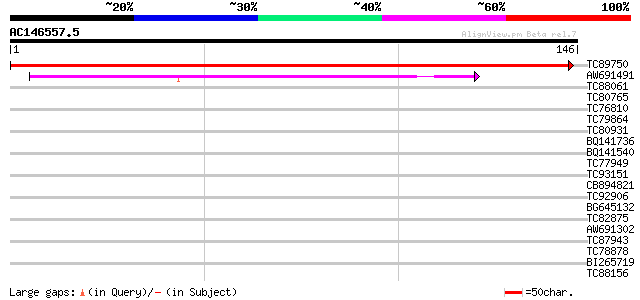
(146 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89750 similar to GP|11558192|emb|CAC17702. transcription facto... 294 8e-81
AW691491 similar to GP|10443853|gb| E2F transcription factor-3 E... 66 6e-12
TC88061 homologue to SP|Q43117|KPYA_RICCO Pyruvate kinase isozym... 33 0.053
TC80765 31 0.16
TC76810 homologue to GP|17645764|emb|CAD19054. unnamed protein p... 29 0.59
TC79864 28 1.0
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 27 2.2
BQ141736 similar to PIR|T06291|T062 extensin homolog T9E8.80 - A... 27 2.2
BQ141540 similar to SP|Q9PJY2|PMPB Probable outer membrane prote... 27 3.8
TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {A... 27 3.8
TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {... 27 3.8
CB894821 similar to GP|4884835|gb|A aminohydroxybenzoic acid syn... 26 5.0
TC92906 similar to GP|22830946|dbj|BAC15811. putative NADH dehyd... 26 5.0
BG645132 similar to GP|4092771|gb|A disease resistance gene homo... 26 6.5
TC82875 similar to GP|15131529|emb|CAC48384. ethylene receptor {... 26 6.5
AW691302 26 6.5
TC87943 similar to GP|21554556|gb|AAM63612.1 unknown {Arabidopsi... 26 6.5
TC78878 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis t... 26 6.5
BI265719 similar to GP|4580456|gb|A unknown protein {Arabidopsis... 25 8.5
TC88156 similar to GP|17132837|dbj|BAB75402. ORF_ID:alr3703~hypo... 25 8.5
>TC89750 similar to GP|11558192|emb|CAC17702. transcription factor (E2F)
{Chenopodium rubrum}, partial (42%)
Length = 1007
Score = 294 bits (753), Expect = 8e-81
Identities = 144/145 (99%), Positives = 145/145 (99%)
Frame = +2
Query: 1 MSGTRTPPATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTP 60
MSGTRTPPATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTP
Sbjct: 113 MSGTRTPPATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTP 292
Query: 61 QLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPG 120
QLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPG
Sbjct: 293 QLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPG 472
Query: 121 SNIGSPSGNNLTPAGPCRYDSSLGI 145
SNIGSPSGNNLTPAGPCRYDSSLG+
Sbjct: 473 SNIGSPSGNNLTPAGPCRYDSSLGL 547
>AW691491 similar to GP|10443853|gb| E2F transcription factor-3 E2F3
{Arabidopsis thaliana}, partial (9%)
Length = 624
Score = 65.9 bits (159), Expect = 6e-12
Identities = 40/117 (34%), Positives = 59/117 (50%), Gaps = 1/117 (0%)
Frame = +2
Query: 6 TPPATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAP-DHRRNQDLETEAIVVKTPQLKR 64
+P T A + +R L F S KPPF +YH FA D R+ D EA++V++P +KR
Sbjct: 152 SPMPTGAQIRPPLKRHLAFVSTKPPFAPTDEYHTFAAVDSRKVADHVNEAVIVRSPVIKR 331
Query: 65 KSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGS 121
K+ + E DS A NS +TP+S K G+ KS + ++ G P +
Sbjct: 332 KNGMNESEGDSQKSSNSPGYNAMNSPFKTPLSAKGGRANKS----RASKEGKSCPSN 490
Score = 33.1 bits (74), Expect = 0.041
Identities = 20/48 (41%), Positives = 25/48 (51%)
Frame = +1
Query: 98 KAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAGPCRYDSSLGI 145
K GKG + + + Q + PS LTPAG CRYDSSLG+
Sbjct: 430 KRGKGKQVKGIQRRQIMSLQHLSQMLVPPSP--LTPAGSCRYDSSLGL 567
>TC88061 homologue to SP|Q43117|KPYA_RICCO Pyruvate kinase isozyme A
chloroplast precursor (EC 2.7.1.40). [Castor bean]
{Ricinus communis}, partial (84%)
Length = 1765
Score = 32.7 bits (73), Expect = 0.053
Identities = 25/94 (26%), Positives = 39/94 (40%), Gaps = 4/94 (4%)
Frame = +1
Query: 2 SGTRTPPATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTPQ 61
+ T TPP + P P S+ +PP + AP H R+ L+ + T
Sbjct: 148 TSTPTPPTSHLPPSTSHSAASPLSAPQPPT*QSPT--TTAPRHHRHHHLQIQTQSKSTQS 321
Query: 62 LKRKSE----AADFEADSGDRMTPGSTAAANSSV 91
K+ S+ AA E +S + P A+ NS +
Sbjct: 322 PKQSSKKTVSAAQEEPNSSALLVPPHAASTNSKL 423
>TC80765
Length = 373
Score = 31.2 bits (69), Expect = 0.16
Identities = 19/62 (30%), Positives = 26/62 (41%)
Frame = +2
Query: 64 RKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNI 123
R+ +A D+ +TP T A + TPV GKA GK+ + P NI
Sbjct: 143 RQDPSATPSGDATPEITPDITPDAATPAATPVDGKAPTDGKAPNLPSSPAPSGAVPTGNI 322
Query: 124 GS 125
S
Sbjct: 323 VS 328
>TC76810 homologue to GP|17645764|emb|CAD19054. unnamed protein product
{Glycine max}, partial (98%)
Length = 1439
Score = 29.3 bits (64), Expect = 0.59
Identities = 20/63 (31%), Positives = 26/63 (40%)
Frame = +2
Query: 83 STAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAGPCRYDSS 142
S A+NS P KAG ++ TK S TPG G P G A P + +
Sbjct: 77 SLMASNSQKDIPSDEKAGSA--ENKNTKDEASSNATPGPGTGFPPGTGF-QANPFDFSAM 247
Query: 143 LGI 145
G+
Sbjct: 248 SGL 256
>TC79864
Length = 1551
Score = 28.5 bits (62), Expect = 1.0
Identities = 19/74 (25%), Positives = 29/74 (38%)
Frame = +1
Query: 63 KRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSN 122
K K + + + +R S + + S P + G+G SSR K ++ P S
Sbjct: 1216 KLKRRVSQLDKNKFERKNTSSFGSGSGSGSRPPRKRGGRGTSSSRPAKSAKTSVY-PSSF 1392
Query: 123 IGSPSGNNLTPAGP 136
NNL P P
Sbjct: 1393 XRLXRXNNLAPVHP 1434
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 27.3 bits (59), Expect = 2.2
Identities = 23/75 (30%), Positives = 32/75 (42%), Gaps = 8/75 (10%)
Frame = +1
Query: 66 SEAADFEADSGDRMTPG--------STAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQ 117
+E+A F + D TP STAAA S+ T + A +S + +G
Sbjct: 619 TESAAFPDSTADSTTPATAAATPSTSTAAAFSAAATAATASAVTATATSTASSAATNGWC 798
Query: 118 TPGSNIGSPSGNNLT 132
GSN+ S S LT
Sbjct: 799 WNGSNLCSRSWGALT 843
>BQ141736 similar to PIR|T06291|T062 extensin homolog T9E8.80 - Arabidopsis
thaliana, partial (4%)
Length = 1099
Score = 27.3 bits (59), Expect = 2.2
Identities = 19/55 (34%), Positives = 25/55 (44%), Gaps = 10/55 (18%)
Frame = +2
Query: 76 GDRMTPG----------STAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPG 120
GDR TPG S A A + + +G KGGK + T+ RSG + G
Sbjct: 473 GDRPTPGREKKNQRTKESGARAPKNQKERAAGGRRKGGKEAGGTETGRSGEREGG 637
>BQ141540 similar to SP|Q9PJY2|PMPB Probable outer membrane protein pmpB
precursor (Polymorphic membrane protein B)., partial
(1%)
Length = 1156
Score = 26.6 bits (57), Expect = 3.8
Identities = 19/66 (28%), Positives = 28/66 (41%), Gaps = 2/66 (3%)
Frame = +2
Query: 72 EADSGDRMTPGSTAAANSSVQTPVSGKA-GKGGKSSRMTKCNRSGTQTPGSNIGSPSGNN 130
E G + PG + P +GK + G KC++ PG + G+P+ NN
Sbjct: 185 EPPPGPKTRPGRVSPHQKG--GPRAGKVKSRWGGGKEKGKCSKGKGDRPGRDRGNPTKNN 358
Query: 131 -LTPAG 135
PAG
Sbjct: 359 GRAPAG 376
>TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {Arabidopsis
thaliana}, partial (34%)
Length = 1297
Score = 26.6 bits (57), Expect = 3.8
Identities = 20/65 (30%), Positives = 27/65 (40%)
Frame = +3
Query: 75 SGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPA 134
+G PG AA S P G AG G + +PGSN +PSGN+ +
Sbjct: 813 AGGPPAPGPGGAATS----PGPGGAGASGPGGAA-----AAPGSPGSNSTAPSGNSGSFV 965
Query: 135 GPCRY 139
P +
Sbjct: 966 APSNF 980
>TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 657
Score = 26.6 bits (57), Expect = 3.8
Identities = 24/83 (28%), Positives = 33/83 (38%), Gaps = 12/83 (14%)
Frame = +3
Query: 76 GDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTP-----GSN------IG 124
G TPG+ SS TP+ G A G ++ T + T GS+ G
Sbjct: 381 GTPSTPGNLFGGASSASTPLFGGAASGSTTASTTMFGGASPATSTAFGFGSSASSTP*FG 560
Query: 125 SPSGNNLTPA-GPCRYDSSLGIA 146
+PS + TP G S G+A
Sbjct: 561 APSSASSTPLFGAPSSTPSFGVA 629
>CB894821 similar to GP|4884835|gb|A aminohydroxybenzoic acid synthase
{Streptomyces collinus}, partial (4%)
Length = 708
Score = 26.2 bits (56), Expect = 5.0
Identities = 24/102 (23%), Positives = 38/102 (36%), Gaps = 4/102 (3%)
Frame = +3
Query: 43 DHRRNQDLETEAIVVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKG 102
DHR + + T + K P AA + +S T A ++ Q
Sbjct: 195 DHRGRR*VHTSSNTSKQPP---HGHAAGWRDESAAPQTTERD*QAGTAAQA--------- 338
Query: 103 GKSSRMTKCNRSGTQTPGSNI----GSPSGNNLTPAGPCRYD 140
+ R+ + GT+TPG+N G +L P P +D
Sbjct: 339 --ADRVRTARQEGTKTPGTNATQKRGGARTTSLAPKAPTSHD 458
>TC92906 similar to GP|22830946|dbj|BAC15811. putative NADH dehydrogenase
{Oryza sativa (japonica cultivar-group)}, partial (35%)
Length = 857
Score = 26.2 bits (56), Expect = 5.0
Identities = 14/47 (29%), Positives = 21/47 (43%)
Frame = -1
Query: 98 KAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAGPCRYDSSLG 144
KAG+ G + C+R Q+P + S G + P+ P LG
Sbjct: 341 KAGRTG----LPVCSRKPAQSPSAKTSSTDGTRIHPSTPILPPGELG 213
>BG645132 similar to GP|4092771|gb|A disease resistance gene homolog 1A
{Brassica napus}, partial (4%)
Length = 790
Score = 25.8 bits (55), Expect = 6.5
Identities = 14/43 (32%), Positives = 25/43 (57%)
Frame = +3
Query: 51 ETEAIVVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQT 93
E + I + +K +SE +F+ TPGS++++NSS +T
Sbjct: 342 EIKNIKISIRGIKERSERYNFQISQ----TPGSSSSSNSSRET 458
>TC82875 similar to GP|15131529|emb|CAC48384. ethylene receptor {Fragaria x
ananassa}, partial (39%)
Length = 1118
Score = 25.8 bits (55), Expect = 6.5
Identities = 11/28 (39%), Positives = 19/28 (67%)
Frame = -3
Query: 14 DQIMQQRQLPFSSMKPPFLAAADYHRFA 41
D +Q +QLP +S P +++A +H+FA
Sbjct: 1071 DLTIQTQQLPRASFTPFYVSALLHHKFA 988
>AW691302
Length = 1028
Score = 25.8 bits (55), Expect = 6.5
Identities = 13/39 (33%), Positives = 19/39 (48%)
Frame = -3
Query: 83 STAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGS 121
+TA SVQTP + GGK + R+G + G+
Sbjct: 417 NTAQLTLSVQTPPRKEVAGGGKPASQYSAARTGNRRTGT 301
>TC87943 similar to GP|21554556|gb|AAM63612.1 unknown {Arabidopsis
thaliana}, partial (53%)
Length = 678
Score = 25.8 bits (55), Expect = 6.5
Identities = 10/30 (33%), Positives = 13/30 (43%)
Frame = -3
Query: 2 SGTRTPPATAAPDQIMQQRQLPFSSMKPPF 31
SG +PD + R +PF PPF
Sbjct: 331 SGVMDASMLTSPDTSLSSRDVPFGRFSPPF 242
>TC78878 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis thaliana},
partial (53%)
Length = 1839
Score = 25.8 bits (55), Expect = 6.5
Identities = 9/31 (29%), Positives = 16/31 (51%)
Frame = -2
Query: 7 PPATAAPDQIMQQRQLPFSSMKPPFLAAADY 37
PP + ++ LP+S KPP +++ Y
Sbjct: 782 PPVSIHSPPSSSRKNLPYSQQKPPIVSSCKY 690
>BI265719 similar to GP|4580456|gb|A unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 660
Score = 25.4 bits (54), Expect = 8.5
Identities = 11/17 (64%), Positives = 13/17 (75%)
Frame = +3
Query: 80 TPGSTAAANSSVQTPVS 96
TP +TA NSS+QTP S
Sbjct: 495 TPPATANNNSSIQTPTS 545
>TC88156 similar to GP|17132837|dbj|BAB75402. ORF_ID:alr3703~hypothetical
protein {Nostoc sp. PCC 7120}, partial (5%)
Length = 673
Score = 25.4 bits (54), Expect = 8.5
Identities = 23/73 (31%), Positives = 33/73 (44%), Gaps = 1/73 (1%)
Frame = -3
Query: 11 AAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTPQLKRKSEAAD 70
AAPDQI QQ Q L + ++ P R+ +D TE I K P+++ + D
Sbjct: 470 AAPDQISQQYQFQQFQQHQQELLQSTLYQQKPKQRQCKD--TEPIP*K-PEVEGRCRGCD 300
Query: 71 -FEADSGDRMTPG 82
E G+R G
Sbjct: 299 RVEGHGGERSQLG 261
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.125 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,728,786
Number of Sequences: 36976
Number of extensions: 42846
Number of successful extensions: 233
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 231
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 232
length of query: 146
length of database: 9,014,727
effective HSP length: 87
effective length of query: 59
effective length of database: 5,797,815
effective search space: 342071085
effective search space used: 342071085
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146557.5


