

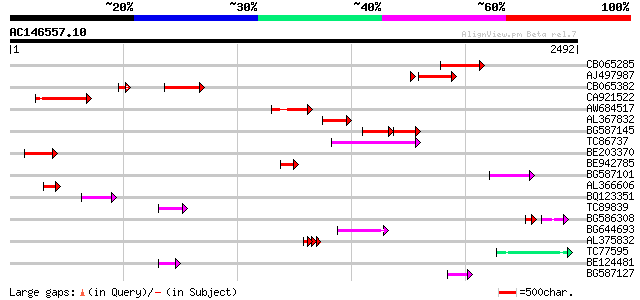
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146557.10 - phase: 0
(2492 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 400 e-111
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 300 5e-81
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 298 2e-80
CA921522 251 2e-66
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 248 2e-65
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 243 7e-64
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 151 6e-59
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 181 4e-45
BE203370 151 3e-36
BE942785 133 1e-30
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 113 1e-24
AL366606 107 6e-23
BQ123351 104 4e-22
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 92 2e-18
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 62 2e-18
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 84 7e-16
AL375832 83 1e-15
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 79 2e-14
BE124481 74 9e-13
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 66 1e-10
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 400 bits (1029), Expect = e-111
Identities = 190/197 (96%), Positives = 194/197 (98%)
Frame = -2
Query: 1891 KSKDCEEPLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNN 1950
KSKDCEEPLI EGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGH+IPFTARILFECTNN
Sbjct: 591 KSKDCEEPLIGEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNN 412
Query: 1951 MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYF 2010
MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHA LIPYRDYARRLLTYF
Sbjct: 411 MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYF 232
Query: 2011 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGE 2070
TKVELHHIPRDENQMADALATLSSMFRVNHWNDVP+IKVQRLERPSHVFAIG+VIDQ GE
Sbjct: 231 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFAIGNVIDQTGE 52
Query: 2071 NVVDYKPWYYDIKQFLL 2087
NVVDYKPWYYDIKQFL+
Sbjct: 51 NVVDYKPWYYDIKQFLV 1
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 300 bits (768), Expect = 5e-81
Identities = 133/167 (79%), Positives = 156/167 (92%)
Frame = -2
Query: 1795 KTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYDIVFKT 1854
KTCCALAWAAKRLRHY++NHTTWL+S+MDPIKYIFEK A+TG+IARWQMLLSEYDI +++
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1855 QKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSKWGLVF 1914
QKAIKGSILADHLA+QPL+DY+PI+FDFPDEEIMYLK KDC+EPL EGPDP+S WGL+F
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 1915 DGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGI 1961
DGAVN YG GIGAV+++P+G HIPFTAR+ F+CTNN+AEYEACI GI
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
Score = 48.1 bits (113), Expect = 4e-05
Identities = 20/24 (83%), Positives = 23/24 (95%)
Frame = +2
Query: 1761 CVLGQQDETGKKEHAIYYLSKKFT 1784
CVLGQQDETG+KEHAIYYLS+K +
Sbjct: 38 CVLGQQDETGRKEHAIYYLSRKLS 109
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 298 bits (762), Expect = 2e-80
Identities = 146/175 (83%), Positives = 153/175 (87%)
Frame = -2
Query: 682 PVQQQQQQQQQQYTRPTFPPIPMLYAELLPTLLQRGHCTTRQGKPPPDPLPPRFRSDLKC 741
PV QQ+Q QQ RPTFP IPMLYAE LPTLL RGHCT RQGKPPPDPLPPRFRSDLKC
Sbjct: 518 PVHQQRQ*QQ---ARPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPPPDPLPPRFRSDLKC 348
Query: 742 DFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPNHAAVNMIEVCEEAP 801
DFHQGALGHDVEGCYALK+IVKKLI+QGKLTFENNVPHVLDNPLPNHAAVNMIEV EEAP
Sbjct: 347 DFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPNHAAVNMIEVYEEAP 168
Query: 802 RLDVRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRDPLGCHAVQDDIQSLMNDN 856
LDVRNV TPLVPLHIKLC+ASLF HDHA C +PLGC VQ+DIQSLMN+N
Sbjct: 167 GLDVRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQNDIQSLMNNN 3
Score = 59.3 bits (142), Expect = 2e-08
Identities = 33/50 (66%), Positives = 38/50 (76%)
Frame = -1
Query: 479 EENTQIRTELASLREELAKAHDAMTALLAAQEQPVPVVSTAANVTPTVTT 528
EEN Q+RTELAS+REELAKA+D MTALLAAQEQ S+ + T TV T
Sbjct: 624 EENAQLRTELASMREELAKANDTMTALLAAQEQ-----SSTGSPTTTVAT 490
>CA921522
Length = 767
Score = 251 bits (641), Expect = 2e-66
Identities = 128/256 (50%), Positives = 167/256 (65%), Gaps = 8/256 (3%)
Frame = +3
Query: 111 KTSDVSNHLITKSHIRGFTSKYLLEQANLSTTRQDTL------EAILALLIYGLILFPNL 164
+T +V HLIT+ GF++ +L E+ TT D + +ILALL+YGL+LFP+L
Sbjct: 3 ETDEVKTHLITRGKFLGFSTDFLYER----TTFFDKMGVAYAFNSILALLVYGLVLFPSL 170
Query: 165 DNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHLPSS-- 222
DNFVD+ AI++F S+NPVPTLL DTY +IH RT GRG ILCC LLYRW SHLP +
Sbjct: 171 DNFVDIKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPR 350
Query: 223 FHDNSENWSYSQRIMALTPNEVVWITPAAQAKEIIIGCGDFLNVPLLGTRGGINYNPELA 282
F N EN +S+R+M+LTP EVVW II CG F NVPLLG GGI+YNP LA
Sbjct: 351 FTTNPENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLA 530
Query: 283 MRQFGFPMKSKPINLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSVKHLGVRSGVAHEA 342
QFG+PM+ KP+++ ++ + TG R+ + AW +RR+ LG ++G +
Sbjct: 531 RHQFGYPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSS 710
Query: 343 YTQWVIDRAEEIGMPY 358
YTQWVIDRA +IGMPY
Sbjct: 711 YTQWVIDRAVQIGMPY 758
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana chromosome
II BAC F26H6; putative retroelement pol polyprotein,
partial (1%)
Length = 488
Score = 248 bits (634), Expect = 2e-65
Identities = 125/180 (69%), Positives = 140/180 (77%)
Frame = +1
Query: 1150 VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNGKLVTIHGEEAYLVSQLLSF 1209
+TFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 1210 SCIEAGSAEGTAFQGLTIEGAEPKEAGAAMASLKDAQRAIQEGQTAGWGKVIRLCENKRK 1269
SAEGTAFQGL++EGAEPK+ GAAMASLKDAQ+A+QEGQ A WGK+I+LCENKRK
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 1270 EGLGFSPSSRVSSGVFHSAGFVNAISEEATGSGLRPVFVTPGGIATDWDAIDIPSIMHVS 1329
EGL FSP+S VS+G FHSAGFVN ++EE RP+FV PGGIA DWDA+D+PSIMHVS
Sbjct: 286 EGLRFSPTSGVSTGTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIMHVS 465
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 243 bits (620), Expect = 7e-64
Identities = 118/128 (92%), Positives = 122/128 (95%)
Frame = -1
Query: 1373 QEKKAIQPHQEEIELVNIGTEENKREIKIGATLEEGVKRKIIQLLREYPDIFAWSYEDMP 1432
QE+KAIQPHQEEIEL+N+GTEENKREIK+GA LEEGVKRKI QLLREY DIFA SYEDMP
Sbjct: 384 QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMP 205
Query: 1433 GLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 1492
GLDP IVEHRIPTKPECPPVR KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI
Sbjct: 204 GLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 25
Query: 1493 VPVPKKDG 1500
VPVPKKDG
Sbjct: 24 VPVPKKDG 1
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 151 bits (381), Expect(2) = 6e-59
Identities = 70/136 (51%), Positives = 100/136 (73%)
Frame = +2
Query: 1552 MSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVK 1611
M P+D EKT+FIT GT+CYKVMPFGL NAG+TYQR + +F D +EVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 1612 STDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIR 1671
S H+ +L + F+ L +Y ++LNP KCTFGV SG+ LG+IV+Q+GIEV+P ++ AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 1672 EMPAPRTEKQVRGFLG 1687
++P+P+ ++V+ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 97.4 bits (241), Expect(2) = 6e-59
Identities = 49/120 (40%), Positives = 76/120 (62%)
Frame = +3
Query: 1687 GRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYLLEPPILVPPVEGR 1746
GR+ ++RFIS T C P +KLL N+ VW+++C+EAF+ +K+YL PP+L P G
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 1747 PLIMYLAVFDESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKR 1806
L +Y+A+ ++ VL ++D +K I+Y SK+ TD ETRY LEK A+ +A++
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQK--PIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 181 bits (458), Expect = 4e-45
Identities = 123/398 (30%), Positives = 200/398 (49%), Gaps = 7/398 (1%)
Frame = +1
Query: 1416 LLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPEC----PPVRQ-KLRRTHPDMALKIKSE 1470
+L E+PD+F +++H IP P+ PP+ L L +K
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 1471 VQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVD 1530
++ +D GF+ A ++ V K G +R CVD+R LN + KD +PLP I +
Sbjct: 523 LEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLR 699
Query: 1531 NTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMT 1590
A ++ F+ +D + ++++++ ED+EKT+F T +G F + V PFGL A AT+QR +
Sbjct: 700 RVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYIN 879
Query: 1591 TLFHDMTHKEVEVYVDDMIVKST-DEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGK 1649
H+ V Y+DD+++ +T ++ H + ++ RL L L+P KC F V + K
Sbjct: 880 KTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVK 1059
Query: 1650 LLGFIVSQ-KGIEVDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFK 1708
+GFI++ KG+ DP K+ AIR+ P + K R FLG NY FI + P+ +
Sbjct: 1060 YVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTR 1239
Query: 1709 LLRKNQPVVWNDECQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVLGQQDE 1768
L RK+ P W E + AF +K+ E P+L + ++G VL Q+D
Sbjct: 1240 LTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDG 1419
Query: 1769 TGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKR 1806
TG H + + S++ + E Y + +K A+ WA R
Sbjct: 1420 TG-AAHPVAFHSQRLSPAEYNYPIHDKELLAV-WACLR 1527
>BE203370
Length = 454
Score = 151 bits (382), Expect = 3e-36
Identities = 79/150 (52%), Positives = 101/150 (66%), Gaps = 2/150 (1%)
Frame = +1
Query: 63 CFTFPDFQLVPTLEAYSNLVGLPIAEKTPFTGPGTSLTPLVIAKDLYLKTSDVSNHLITK 122
CFTFPD+QL PTLE YS LVG P+ +K PFTG IA L+L+TS + L K
Sbjct: 4 CFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETSLIKAKLTIK 183
Query: 123 SHIRGFTSKYLLEQAN--LSTTRQDTLEAILALLIYGLILFPNLDNFVDMNAIEVFHSKN 180
++ +K+L +QA+ D +ILALLIYGL+LFPN+DN+VD++AI++F +KN
Sbjct: 184 GNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLTKN 363
Query: 181 PVPTLLADTYHAIHDRTLKGRGYILCCISL 210
PVPTLLAD YH+IHDR GRG IL C L
Sbjct: 364 PVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>BE942785
Length = 460
Score = 133 bits (334), Expect = 1e-30
Identities = 64/82 (78%), Positives = 72/82 (87%)
Frame = -2
Query: 1189 NGKLVTIHGEEAYLVSQLLSFSCIEAGSAEGTAFQGLTIEGAEPKEAGAAMASLKDAQRA 1248
+G LVTIHGEEAYL+SQL SFSCIEAGSAEGTAFQGLT+EG EPK G AMASLKDAQRA
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 1249 IQEGQTAGWGKVIRLCENKRKE 1270
+QE Q AGWG++I+L ENK K+
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 113 bits (282), Expect = 1e-24
Identities = 62/196 (31%), Positives = 97/196 (48%)
Frame = +2
Query: 2109 RFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWM 2168
R+L D LYK+ D + +RCV E E ++ H + H K+ +AG++W
Sbjct: 17 RYLWDEPFLYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWP 196
Query: 2169 AMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPFSMWGIDMIGRIEPKASNGHRF 2228
M D + + KC CQ + N + F +WGID +G +N ++
Sbjct: 197 TMFKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNN--KY 370
Query: 2229 ILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALC 2288
ILVA+DY +KWVEA + VV K K+ I R+GVP +I+D G++ N V + L
Sbjct: 371 ILVAVDYVSKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLL 550
Query: 2289 EEFKIEHHNSSPYRPQ 2304
++ + H ++ Y PQ
Sbjct: 551 KKNGVRHKVATAYHPQ 598
>AL366606
Length = 393
Score = 107 bits (267), Expect = 6e-23
Identities = 58/78 (74%), Positives = 59/78 (75%), Gaps = 6/78 (7%)
Frame = -1
Query: 149 AILALLIYGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCI 208
AI L GLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYIL
Sbjct: 381 AIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYILWLH 202
Query: 209 SLL------YRWFISHLP 220
L +R F S LP
Sbjct: 201 ILFSNRVVYFRTFRSSLP 148
>BQ123351
Length = 725
Score = 104 bits (260), Expect = 4e-22
Identities = 59/153 (38%), Positives = 92/153 (59%)
Frame = +2
Query: 317 FMDAWSKVRRKSVKHLGVRSGVAHEAYTQWVIDRAEEIGMPYPAMRYVSSSTPSMPLPLL 376
F+ AW V ++ LG RS + +E+YT+WVIDRA + G+P+P R SS+T S LP+
Sbjct: 8 FIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLPMT 187
Query: 377 PVTQDMYQEHLAMESCEKQVWKARYNQAENLIMTLDGRDEQKTHENLMLKKELAKARREL 436
T++ Q LA + EK W+ RY +AE+ I TL G+ EQK HE L +++++ + L
Sbjct: 188 SKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDDLL 367
Query: 437 EEKDELLMRDSKRARGRRDFFASYCDSDSESDD 469
+EKD+LL + + + R D + DS+ +D
Sbjct: 368 QEKDKLLKKHITK-KQRMDSMDLFDGPDSDFED 463
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 92.4 bits (228), Expect = 2e-18
Identities = 54/132 (40%), Positives = 68/132 (50%), Gaps = 3/132 (2%)
Frame = +3
Query: 653 PPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQQ---QQQQYTRPTFPPIPMLYAEL 709
PP Q P P QP P Q +Q +P Q Q+ Q Q PIP+ YA+L
Sbjct: 228 PPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADL 407
Query: 710 LPTLLQRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQG 769
LP LL++ T P+ LPP +R DL C FHQGA GHD E CY LK V+KLI+
Sbjct: 408 LPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENN 587
Query: 770 KLTFENNVPHVL 781
+F++ VL
Sbjct: 588 VWSFDDQDIKVL 623
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 62.4 bits (150), Expect(2) = 2e-18
Identities = 39/122 (31%), Positives = 69/122 (55%), Gaps = 4/122 (3%)
Frame = -3
Query: 2338 LHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSL---RVIMEAKLSEAEWCQSRYDQL 2394
L +RT R +T +TPFS+ + +EA+ P EV + SL R+ +L+ ++ L
Sbjct: 462 LWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNN----DRLFNAL 295
Query: 2395 NLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQPD-PRGKWAPNY 2453
IEE+R A+ R Q+YQ ++++ ++K V+ + K+G++VL + + GK N+
Sbjct: 294 ETIEERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNW 115
Query: 2454 EG 2455
EG
Sbjct: 114 EG 109
Score = 50.4 bits (119), Expect(2) = 2e-18
Identities = 22/52 (42%), Positives = 35/52 (67%)
Frame = -2
Query: 2265 YGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI 2316
+G+P +I+TDNG++ +N + CE ++I + +SP PQ NG EA+NK I
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII 530
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 84.0 bits (206), Expect = 7e-16
Identities = 65/230 (28%), Positives = 108/230 (46%), Gaps = 5/230 (2%)
Frame = +2
Query: 1439 VEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKK 1498
++ I P P+ R +P +K +++ ++ GF+ YP V ++ + KK
Sbjct: 35 IDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKK 211
Query: 1499 DGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDRE 1558
DG +RM +D+ LN + K +PLP ID L DN SK F +D G +Q ++ ED
Sbjct: 212 DGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVP 391
Query: 1559 KTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTDEEQH 1618
KT+F +G + VM FG N + M +F D V V+ +D+++ S +E +H
Sbjct: 392 KTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEH 571
Query: 1619 VEYLTKMFERLRKYKLRLNPNKCTFG-----VRSGKLLGFIVSQKGIEVD 1663
+L + L+ L C V G ++S +G++VD
Sbjct: 572 ENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>AL375832
Length = 535
Score = 83.2 bits (204), Expect = 1e-15
Identities = 39/41 (95%), Positives = 40/41 (97%)
Frame = +3
Query: 1290 FVNAISEEATGSGLRPVFVTPGGIATDWDAIDIPSIMHVSE 1330
FVNAISEEATGSGLRP FVTPGGIA+DWDAIDIPSIMHVSE
Sbjct: 3 FVNAISEEATGSGLRPAFVTPGGIASDWDAIDIPSIMHVSE 125
Score = 34.7 bits (78), Expect(2) = 1e-04
Identities = 14/22 (63%), Positives = 16/22 (72%)
Frame = +1
Query: 1328 VSELNHNKPVEHSNPTVPPSFD 1349
+ LNHNKPVEHSN P+FD
Sbjct: 418 ICRLNHNKPVEHSNSMPLPNFD 483
Score = 31.2 bits (69), Expect(2) = 1e-04
Identities = 13/17 (76%), Positives = 13/17 (76%)
Frame = +3
Query: 1350 FPVYEAKDEEGDGIPYE 1366
FPVYE K EGD IPYE
Sbjct: 483 FPVYEPKMREGDDIPYE 533
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 79.0 bits (193), Expect = 2e-14
Identities = 75/353 (21%), Positives = 141/353 (39%), Gaps = 15/353 (4%)
Frame = +2
Query: 2137 QLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPH 2196
+L+ + HD T H G + +++ ++W ++ R C C IH+
Sbjct: 176 KLVQESHDSTAAGHP-GRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVC----GGIHIWRQ 340
Query: 2197 ALNVMSSPWPF-----SMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQ 2251
A P P S +D I + P G +++ V +D +K V + +
Sbjct: 341 AKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEAE 520
Query: 2252 VVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEA 2311
A+ + +G+P I++D G+N + C + S+ Y PQ +G E
Sbjct: 521 ACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTER 700
Query: 2312 ANKNIKRIVQKMVTTYKD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEI 2370
N+ I+ +++ V +D W ++LP R SS GATPF + +G V P+
Sbjct: 701 WNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYH-VDPIPTVE 877
Query: 2371 PSLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKV 2430
+ V+ E + + + D I+ + + A R ++ + + D+ ++V
Sbjct: 878 DTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEASANKRRCPADR------YQV 1039
Query: 2431 GELV---LKRRISQQPDPRGKW------APNYEGPYVVKKAFSGGALILTHMD 2474
G+ V + S +P + W + P+VV+ G H+D
Sbjct: 1040 GDKVWLNVSNYKSPRPSKKLDWLHHKYEVTRFVTPHVVELNVPGTVYPKFHVD 1198
>BE124481
Length = 538
Score = 73.6 bits (179), Expect = 9e-13
Identities = 42/102 (41%), Positives = 51/102 (49%), Gaps = 3/102 (2%)
Frame = +3
Query: 653 PPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQQ---QQQQYTRPTFPPIPMLYAEL 709
PP Q P P QP P Q +Q +P Q Q+ Q Q PIP+ YA+L
Sbjct: 231 PPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADL 410
Query: 710 LPTLLQRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHD 751
LP LL++ T P+ LPP +R DL C FHQGA GHD
Sbjct: 411 LPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHD 536
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 66.2 bits (160), Expect = 1e-10
Identities = 38/109 (34%), Positives = 55/109 (49%)
Frame = +3
Query: 1926 GAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQI 1985
G + SP + + R+ F +NN YEA I G+ A ++I+++ Y DS LV +Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 1986 KGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSS 2034
GE+E + Y ++L L IPR EN ADALA L+S
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALAS 329
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,923,733
Number of Sequences: 36976
Number of extensions: 1115979
Number of successful extensions: 7380
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 6127
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6992
length of query: 2492
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2380
effective length of database: 4,873,415
effective search space: 11598727700
effective search space used: 11598727700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146557.10


