

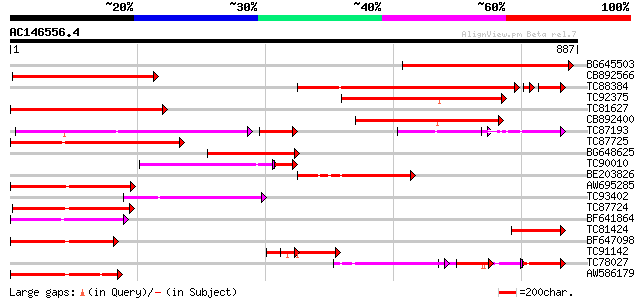
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146556.4 + phase: 0
(887 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG645503 homologue to PIR|T16899|T168 hypothetical protein T19D7... 529 e-150
CB892566 weakly similar to GP|15289776|db Lycopersicon esculentu... 451 e-127
TC88384 homologue to GP|21626661|gb|AAF47055.2 CG13560-PA {Droso... 352 e-110
TC92375 352 3e-97
TC81627 weakly similar to GP|18652501|gb|AAL77135.1 Putative NBS... 310 2e-84
CB892400 297 1e-80
TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Or... 228 1e-68
TC87725 similar to GP|5817341|gb|AAD52714.1| putative NBS-LRR ty... 237 1e-62
BG648625 weakly similar to GP|20385440|gb| resistance gene analo... 224 1e-58
TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR ty... 169 4e-45
BE203826 174 1e-43
AW695285 similar to GP|15487944|gb| NBS/LRR resistance protein-l... 172 4e-43
TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type put... 163 3e-40
TC87724 160 3e-39
BF641864 156 4e-38
TC81424 149 6e-36
BF647098 weakly similar to GP|22324956|gb| putative disease resi... 144 1e-34
TC91142 weakly similar to PIR|T02213|T02213 NBS-LRR type resista... 143 3e-34
TC78027 similar to PIR|D86327|D86327 protein F18O14.17 [imported... 104 3e-34
AW586179 143 3e-34
>BG645503 homologue to PIR|T16899|T168 hypothetical protein T19D7.6 -
Caenorhabditis elegans, partial (9%)
Length = 819
Score = 529 bits (1363), Expect = e-150
Identities = 266/268 (99%), Positives = 266/268 (99%)
Frame = +3
Query: 615 VDGVEMLPQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLK 674
VDGVEMLPQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLK
Sbjct: 3 VDGVEMLPQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLK 182
Query: 675 SLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFK 734
SLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFK
Sbjct: 183 SLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFK 362
Query: 735 SLSDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKL 794
SLSDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKL
Sbjct: 363 SLSDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKL 542
Query: 795 RLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRC 854
RLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRC
Sbjct: 543 RLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRC 722
Query: 855 KRGIGEDWHKIAHIPISPSSVETTTXNK 882
KRGIGEDWHKIAHIPISPSSVETTT K
Sbjct: 723 KRGIGEDWHKIAHIPISPSSVETTTPTK 806
>CB892566 weakly similar to GP|15289776|db Lycopersicon esculentum resistance
complex protein I2C-2 like protein, partial (1%)
Length = 688
Score = 451 bits (1160), Expect = e-127
Identities = 229/229 (100%), Positives = 229/229 (100%)
Frame = +2
Query: 5 LLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKEWLQQ 64
LLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKEWLQQ
Sbjct: 2 LLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKEWLQQ 181
Query: 65 LRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDIAEER 124
LRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDIAEER
Sbjct: 182 LRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDIAEER 361
Query: 125 MKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELSVYSI 184
MKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELSVYSI
Sbjct: 362 MKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELSVYSI 541
Query: 185 VGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIEN 233
VGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIEN
Sbjct: 542 VGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIEN 688
>TC88384 homologue to GP|21626661|gb|AAF47055.2 CG13560-PA {Drosophila
melanogaster}, partial (3%)
Length = 1570
Score = 352 bits (904), Expect(3) = e-110
Identities = 194/353 (54%), Positives = 240/353 (67%), Gaps = 5/353 (1%)
Frame = -3
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL++LQLGG+LHIK LENVS+E DA+E NLI KK+L+RLYLSWG+ NSQ D E+VL
Sbjct: 1364 ELNDLQLGGRLHIKGLENVSSEWDAKEANLIGKKELNRLYLSWGSHANSQGIDTDVEQVL 1185
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EALEPH+ GLK FG+ GY G FP WM+N SIL+GLV I YNC NC+ LPP GKLPCLT
Sbjct: 1184 EALEPHT-GLKGFGIEGYVGIHFPHWMRNASILEGLVDITFYNCNNCQRLPPLGKLPCLT 1008
Query: 570 ILYLSGMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLDI 629
LY+ GMR +KYIDDD+YE +++AF SLK L+L LPNLER+L+ +GVEMLPQL +I
Sbjct: 1007 TLYVFGMRDLKYIDDDIYESTSKRAFISLKNLTLLGLPNLERMLKAEGVEMLPQLSYFNI 828
Query: 630 TNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSED---VAGNNLKSLSISKFANLKE 686
+NVPKL L SL S+E L L + + + + +NLK L I F LK
Sbjct: 827 SNVPKLALPSLPSIELLDVGENKYRFLPQYKVVSLFPERIVCSMHNLKLLIIGNFHKLKV 648
Query: 687 LPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLTCL 746
LP +L L+ LE L I RC+E+ESFS H L+G+ SLR +++ C SLS+GM L L
Sbjct: 647 LPDDLHCLSVLEELHISRCDELESFSMHALQGMISLRVLTIDLCGKLISLSEGMGDLASL 468
Query: 747 ETLHIYYCPQLVFPHNMNSLASLRQLLLVEC--NESILDGIEGIPSLQKLRLF 797
E L I+ C QLV P MN L SLRQ+ + N IL G+E IPSLQ L LF
Sbjct: 467 ERLVIHGCSQLVLPSKMNKLTSLRQVNISHSGRNNRILQGLEVIPSLQNLTLF 309
Score = 65.9 bits (159), Expect(3) = e-110
Identities = 29/42 (69%), Positives = 33/42 (78%)
Frame = -2
Query: 828 SLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
SLP++FQ L NL TL I GC LEKRCK+G GEDW KIAH+P
Sbjct: 225 SLPNSFQNLINLHTLLIVGCSKLEKRCKKGTGEDWQKIAHVP 100
Score = 23.1 bits (48), Expect(3) = e-110
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = -1
Query: 805 LPDWLGAMTSLQVLAI 820
LP+ LGAMTSLQ + I
Sbjct: 295 LPESLGAMTSLQRVEI 248
>TC92375
Length = 837
Score = 352 bits (904), Expect = 3e-97
Identities = 184/264 (69%), Positives = 207/264 (77%), Gaps = 7/264 (2%)
Frame = +3
Query: 520 KHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGMRYI 579
K G+ GY G FP WM+NTSILKGLVSIILY+CKNCR +PPFGKLPCLT L +S MR +
Sbjct: 42 KECGLQGYMGAHFPHWMRNTSILKGLVSIILYDCKNCRQIPPFGKLPCLTFLSVSRMRDL 221
Query: 580 KYIDDDLYEPETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLDITNVPKLTLTS 639
KYIDD LYEP TEKAFTSLKK +L DLPNLERVL+ +GVEML QLL LDIT+VPKL L S
Sbjct: 222 KYIDDSLYEPTTEKAFTSLKKFTLADLPNLERVLKAEGVEMLQQLLKLDITDVPKLALQS 401
Query: 640 LLSVESLSASGGNEELLKSFFYNNCSEDVAG-------NNLKSLSISKFANLKELPVELG 692
L S+ESL AS GNEELLKS FYNNC+E+VA NNLKSL ISKFA L ELPVEL
Sbjct: 402 LPSMESLYASRGNEELLKSIFYNNCNEEVASSSREISCNNLKSLCISKFAKLMELPVELS 581
Query: 693 PLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLTCLETLHIY 752
L AL++L I C+EMES SEHLL+GL SLR +++ C FKSLS+GMRHLTCLETL IY
Sbjct: 582 RLGALDNLKIAHCDEMESLSEHLLQGLRSLRTLAIHECGRFKSLSNGMRHLTCLETLEIY 761
Query: 753 YCPQLVFPHNMNSLASLRQLLLVE 776
CPQLV NMNSL S R+L+L E
Sbjct: 762 NCPQLVSLDNMNSLTSPRRLVLWE 833
>TC81627 weakly similar to GP|18652501|gb|AAL77135.1 Putative NBS-LRR type
resistance protein {Oryza sativa}, partial (6%)
Length = 890
Score = 310 bits (793), Expect = 2e-84
Identities = 168/248 (67%), Positives = 199/248 (79%), Gaps = 1/248 (0%)
Frame = +2
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
M D LLGI+I+NL SFV+EEL+T+LGVGELTQ L LT IRAVL+DAE+KQIT+ AVK+
Sbjct: 146 MTDVLLGIVIENLISFVREELSTFLGVGELTQKLCGNLTAIRAVLQDAEEKQITSRAVKD 325
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L DAA+VLDDILD+CS T KAHG+NK I RFHP KIL +R IGKRMKE+AK+ID+I
Sbjct: 326 WLQKLADAAHVLDDILDDCSTTSKAHGDNKWIARFHPKKILAQRAIGKRMKEVAKKIDEI 505
Query: 121 AEERMKFGLHVGVI-ERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEEL 179
AE R+K+GL V V E Q D+ RQTTSVITE KVYGRD D E IVE L+ HA DSEEL
Sbjct: 506 AEGRIKYGLQVKVTEEHQRGDDEWRQTTSVITEPKVYGRDHDIEKIVEVLVSHAIDSEEL 685
Query: 180 SVYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNP 239
SVYSIVG GG GKTTLAQ VFNDER+ T ++ + +S + MK+LESII++T+GKNP
Sbjct: 686 SVYSIVGVGGLGKTTLAQVVFNDERLDTLYERLTNMKLSSSSSMMKILESIIKSTVGKNP 865
Query: 240 HLSSLESM 247
LSSLESM
Sbjct: 866 *LSSLESM 889
>CB892400
Length = 738
Score = 297 bits (761), Expect = 1e-80
Identities = 161/241 (66%), Positives = 183/241 (75%), Gaps = 10/241 (4%)
Frame = +3
Query: 542 LKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKAFTSLKKL 601
LK LV IILY+CKNC+ L P GKLPCLT L+LSGMR +KYI+DDLYEP +EKAF SLKKL
Sbjct: 3 LKSLVCIILYDCKNCKQLSPLGKLPCLTTLFLSGMRNLKYIEDDLYEPASEKAFASLKKL 182
Query: 602 SLHDLPNLERVLEVDGVEMLPQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFY 661
+L +LPNLERVLEV+GVEMLPQL L I VPKL L+SL SVE L +GGNEELLKS F
Sbjct: 183 TLRNLPNLERVLEVEGVEMLPQLSFLSIECVPKLALSSLPSVEFLFVTGGNEELLKSIFN 362
Query: 662 NNCSED-------VAGN---NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESF 711
NNC+ED AGN NLK L IS FA LKELP ELG L+ LE L I+ C+E+ESF
Sbjct: 363 NNCNEDATLSPQGTAGNNIYNLKLLYISDFAKLKELPDELGTLSTLEVLHIQCCDEIESF 542
Query: 712 SEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQ 771
SEHLL+GLSSLR + + SC FKSLS+ RHLTCLE L I CPQ VF +NMNSL SL +
Sbjct: 543 SEHLLQGLSSLRTLCIHSCHEFKSLSEVTRHLTCLEGLEIIDCPQCVFSNNMNSLTSLHR 722
Query: 772 L 772
L
Sbjct: 723 L 725
>TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 3119
Score = 228 bits (582), Expect(2) = 1e-68
Identities = 136/381 (35%), Positives = 210/381 (54%), Gaps = 9/381 (2%)
Frame = +3
Query: 9 LIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKEWLQQLRDA 68
L+ L S E+ GV + + L + + I+AVL DAE KQ + AV+ W+++L D
Sbjct: 114 LVNRLASAAFREIGRIYGVMDELERLKKTVESIKAVLLDAEDKQEQSHAVQLWVRRLNDL 293
Query: 69 AYVLDDILDECSITL------KAHGNNKR--ITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
DD++DE I KAH N + F P + RR + +++I + D+
Sbjct: 294 LLPADDLIDEFFIEDMIHKRDKAHKNKVKQVFHSFSPSRTAFRRKMAHEIEKIQRSFKDV 473
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
++ L+ V+ + RR+T S + ES++ GR+ DK I+ LLR + + + +S
Sbjct: 474 EKDMSYLKLNSNVVVVAKTNNVRRETCSYVLESEIIGREDDKNKIIS-LLRQSHEHQNVS 650
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
+ +IVG GG GKT LAQ V+ D VK F+ +WVCVS + + +L++++ + ++
Sbjct: 651 LVAIVGIGGLGKTALAQLVYKDGEVKNLFEKHMWVCVSDNFDFKTILKNMVASLTKEDVV 830
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
+L+ +Q +Q L RYLLVLDDVW E EKW++L+ L+ G +G+ +++TT IV
Sbjct: 831 NKTLQELQSMLQVNLTGKRYLLVLDDVWNESFEKWDQLRPYLMCGAQGSKVVMTTCSKIV 1010
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGE-NREERAELVAIGKKLVRKCVGSPLAAKV 359
A MG SD H L L+ + W LFK FG+ L +IGKK+ KC G PLA +
Sbjct: 1011ADRMGVSDQHVLRGLTPEKSWVLFKNIVFGDVTVGVNQPLESIGKKIAEKCKGVPLAIRS 1190
Query: 360 LGSSLCCTSNEHQWISVLESE 380
LG L S E +WI+VL+ E
Sbjct: 1191LGGILRSESKESEWINVLQGE 1253
Score = 66.6 bits (161), Expect = 4e-11
Identities = 49/134 (36%), Positives = 72/134 (53%), Gaps = 2/134 (1%)
Frame = +1
Query: 738 DGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKLRLF 797
D +++LT LE L +++SL+S +Q ++E D + +PSLQK+++
Sbjct: 2608 DWLKNLTSLENL------------DLDSLSS-QQFEVIEM--WFKDDLICLPSLQKIQIR 2742
Query: 798 NFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPI--LEKRCK 855
+K+LPDW+ ++SLQ LA+ L LP+ +L NL TL I GCPI
Sbjct: 2743 TC-RLKALPDWICNISSLQHLAVYGCESLVDLPEGMPRLTNLHTLEIIGCPISYYYDEFL 2919
Query: 856 RGIGEDWHKIAHIP 869
R G W KIAHIP
Sbjct: 2920 RKTGATWSKIAHIP 2961
Score = 51.2 bits (121), Expect(2) = 1e-68
Identities = 24/61 (39%), Positives = 38/61 (61%), Gaps = 1/61 (1%)
Frame = +1
Query: 391 MSALRISYFNLKLSLRPCFAFCAVFPKGFEMVKENLIHLWMANGLVTSRGNLQ-MEHVGD 449
+ L++SY NL R CFA+C++FP+ +E K+ LI +WMA G + Q M+ VG+
Sbjct: 1291 LPVLKLSYQNLSPQQRQCFAYCSLFPQDWEFEKDELIQMWMAQGYLGCSVEKQCMKDVGN 1470
Query: 450 E 450
+
Sbjct: 1471 Q 1473
Score = 42.4 bits (98), Expect = 8e-04
Identities = 42/154 (27%), Positives = 76/154 (49%), Gaps = 5/154 (3%)
Frame = +1
Query: 607 PNLERVLEV--DGVEMLPQLLNLDIT--NVPKLTLTSLLSVESLSASGGNEELLKSFFYN 662
PN+++ LE+ E+L LN+ + ++ L+ L S + G E + K + N
Sbjct: 2443 PNIKKKLELWSCSAEILEATLNIAESQYSIGFPPLSILKSFHIIETIMGMENVPKDWLKN 2622
Query: 663 NCSEDVAGNNLKSLSISKFANLKE-LPVELGPLTALESLSIERCNEMESFSEHLLKGLSS 721
S + +L SLS +F ++ +L L +L+ + I C +++ + + +SS
Sbjct: 2623 LTS--LENLDLDSLSSQQFEVIEMWFKDDLICLPSLQKIQIRTCR-LKALPDWICN-ISS 2790
Query: 722 LRNMSVFSCSGFKSLSDGMRHLTCLETLHIYYCP 755
L++++V+ C L +GM LT L TL I CP
Sbjct: 2791 LQHLAVYGCESLVDLPEGMPRLTNLHTLEIIGCP 2892
Score = 42.0 bits (97), Expect = 0.001
Identities = 23/75 (30%), Positives = 40/75 (52%), Gaps = 3/75 (4%)
Frame = +2
Query: 517 SGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTIL---YL 573
SG + + G +W+ S + ++ + LYNC++ R+LPP +LP L L +L
Sbjct: 2045 SGFRKLSIQQPKGLSLSNWL---SPVTNIIELCLYNCQDLRYLPPLERLPFLKSLDLRWL 2215
Query: 574 SGMRYIKYIDDDLYE 588
+ + YI Y + L+E
Sbjct: 2216 NQLEYIYYEEPILHE 2260
Score = 35.0 bits (79), Expect = 0.13
Identities = 44/184 (23%), Positives = 87/184 (46%), Gaps = 5/184 (2%)
Frame = +2
Query: 666 EDVAGNNLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEH---LLKGLSSL 722
E + + L++L + + + K E+ L+ L R +MES S ++ L L
Sbjct: 1691 ESLDSSRLRTLIVMSYNDYKFRRPEISVFRNLKYL---RFLKMESCSRQGVGFIEKLKHL 1861
Query: 723 RNMSVFSCSGFKSLSDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLL--VECNES 780
R++ + + +SLS + +L CL+T+ + + P ++ L +LR L + V +
Sbjct: 1862 RHLDLRNYDSGESLSKSIGNLVCLQTIKL-NDSVVDSPEVVSKLINLRHLTINHVTFKDK 2038
Query: 781 ILDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQ 840
G + S+Q+ P SL +WL +T++ L + + +L LP ++L L+
Sbjct: 2039 TPSGFRKL-SIQQ------PKGLSLSNWLSPVTNIIELCLYNCQDLRYLPP-LERLPFLK 2194
Query: 841 TLTI 844
+L +
Sbjct: 2195 SLDL 2206
>TC87725 similar to GP|5817341|gb|AAD52714.1| putative NBS-LRR type disease
resistance protein {Pisum sativum}, partial (44%)
Length = 899
Score = 237 bits (605), Expect = 1e-62
Identities = 136/276 (49%), Positives = 174/276 (62%), Gaps = 3/276 (1%)
Frame = +3
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MADALLG++ +NL + Q E +T G+ Q LS L I+AVL+DAEKKQ ++K
Sbjct: 69 MADALLGVVFENLTALHQNEFSTISGIKSKAQKLSDNLVHIKAVLEDAEKKQFKELSIKL 248
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ L+DA YVLDDILDE SI G + T P I R IG R+KEI + +D+I
Sbjct: 249 WLQDLKDAVYVLDDILDEYSIK---SGQLRGSTSLKPKNIKFRNEIGNRLKEITRRLDNI 419
Query: 121 AEERMKFGLHVGVIERQPEDEGR--RQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEE 178
AE + KF L +G R+ D+ RQT S+I E KV+GR+ DKE IVEFLL A DS+
Sbjct: 420 AESKNKFSLQMGGTLREIPDQVAEGRQTGSIIAEPKVFGREVDKEKIVEFLLTQAKDSDF 599
Query: 179 LSVYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIEN-TIGK 237
+SVY I G GG GKTTL Q +FND RV HFD K+WVCVS + ++L SI E+ T+ K
Sbjct: 600 ISVYPIFGLGGIGKTTLVQLIFNDVRVSGHFDKKVWVCVSETFSVKRILCSIFESITLXK 779
Query: 238 NPHLSSLESMQQKVQEILQKNRYLLVLDDVWTEDKE 273
P M+ KVQ + Q RYLL LDDVW ++++
Sbjct: 780 CPDF-EYAVMEGKVQGLXQGKRYLLXLDDVWNQNEQ 884
>BG648625 weakly similar to GP|20385440|gb| resistance gene analog {Vitis
vinifera}, partial (21%)
Length = 521
Score = 224 bits (571), Expect = 1e-58
Identities = 113/144 (78%), Positives = 122/144 (84%)
Frame = +3
Query: 310 HHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVLGSSLCCTSN 369
H LA LSDDDI SLFKQ AFG NRE RAELV IG+KLVRKCVGSPLAAKVLGS L S+
Sbjct: 3 HPLAQLSDDDI*SLFKQHAFGANREGRAELVEIGQKLVRKCVGSPLAAKVLGSLLRFKSD 182
Query: 370 EHQWISVLESEFWNLPEVDSIMSALRISYFNLKLSLRPCFAFCAVFPKGFEMVKENLIHL 429
EHQW SV+ESEFWNL + + +MSALR+SYFNLKLSLRPCF FCAVFPK FEM KE I L
Sbjct: 183 EHQWTSVVESEFWNLADDNHVMSALRLSYFNLKLSLRPCFTFCAVFPKDFEMEKEFFIQL 362
Query: 430 WMANGLVTSRGNLQMEHVGDELHN 453
WMANGLVTSRGNLQMEHVG+E+ N
Sbjct: 363 WMANGLVTSRGNLQMEHVGNEVWN 434
>TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR type disease
resistance protein {Pisum sativum}, partial (88%)
Length = 1107
Score = 169 bits (428), Expect(2) = 4e-45
Identities = 84/219 (38%), Positives = 132/219 (59%), Gaps = 3/219 (1%)
Frame = +3
Query: 203 ERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPHLSSLESMQQKVQEILQKNRYLL 262
+ V+ HFDLK WVCVS D + M+V +S++E+ +L+ ++ ++++I ++ R+L
Sbjct: 9 KEVQQHFDLKAWVCVSEDFDIMRVTKSLLESVTSTTWDSKNLDVLRVELKKISREKRFLF 188
Query: 263 VLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIVASIMGTSDAHHLASLSDDDIWS 322
V DD+W ++ WN+L S +NGK G+ ++ITTR VA + T H L LS++D WS
Sbjct: 189 VFDDLWNDNYNDWNELASPFINGKPGSMVIITTRQQKVAEVAHTFPIHKLKLLSNEDCWS 368
Query: 323 LFKQQAFGENREERAELVAI---GKKLVRKCVGSPLAAKVLGSSLCCTSNEHQWISVLES 379
L + A G + + A+ G+K+ RKC G P+AAK +G L + +W S+L S
Sbjct: 369 LLSKHALGSDEFHHSSNTALEETGRKIARKCGGLPIAAKTIGGLLRSKVDITEWTSILNS 548
Query: 380 EFWNLPEVDSIMSALRISYFNLKLSLRPCFAFCAVFPKG 418
WNL D+I+ AL +SY L L+ CFA+C++F KG
Sbjct: 549 NIWNLRN-DNILPALHLSYQYLPSHLKRCFAYCSIFSKG 662
Score = 31.6 bits (70), Expect(2) = 4e-45
Identities = 15/37 (40%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Frame = +1
Query: 415 FPKGFEMVKENLIHLWMANG-LVTSRGNLQMEHVGDE 450
FPK + + ++ L+ LWMA G L S+ +ME +GD+
Sbjct: 652 FPKDYPLDRKELVLLWMAEGFLDCSQRGKKMEELGDD 762
>BE203826
Length = 634
Score = 174 bits (442), Expect = 1e-43
Identities = 98/185 (52%), Positives = 121/185 (64%)
Frame = +2
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
+L NLQ G LHIK L+N+ N D RE NL S + L+RL+L+W +TNS + AE VL
Sbjct: 89 DLPNLQPGDNLHIKGLQNLRNGGDVREPNLSSMR-LNRLHLAWDRNTNS---TNSAEEVL 256
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
AL PH L F ++GY G P+WM + SIL LV + L NC NC LPP GKLP L
Sbjct: 257 GALRPHRD-LTGFRLSGYRGMNIPNWMTDISILGRLVDVKLMNCINCSQLPPLGKLPFLN 433
Query: 570 ILYLSGMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLDI 629
LYLS M +KYIDD YE TE AF SL +++L DLPNLERVL ++GVEML QL L I
Sbjct: 434 TLYLSQMTNVKYIDDSPYEISTENAFPSLTEMTLFDLPNLERVLRIEGVEMLSQLSKLSI 613
Query: 630 TNVPK 634
++P+
Sbjct: 614 QSIPQ 628
>AW695285 similar to GP|15487944|gb| NBS/LRR resistance protein-like protein
{Theobroma cacao}, partial (9%)
Length = 618
Score = 172 bits (437), Expect = 4e-43
Identities = 98/200 (49%), Positives = 126/200 (63%), Gaps = 3/200 (1%)
Frame = +2
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+ALL + + S +Q E +T G+ ++LS L I AVL DAEK+Q+ + +K
Sbjct: 23 MAEALLRAAFEKVNSLLQSEFSTISGIKSKAKNLSTSLNHIEAVLVDAEKRQVKDSYIKV 202
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQQL+DA YVLDDILDECSI G + F+P I+ RR IG R+KEI + +DDI
Sbjct: 203 WLQQLKDAVYVLDDILDECSIESARLGGS---FSFNPKNIVFRRQIGNRLKEITRRLDDI 373
Query: 121 AEERMKFGLHVGVI---ERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSE 177
A+ + KF L G + E E + RQ S+I + +V+GR DKE I EFLL HA DS+
Sbjct: 374 ADIKNKFLLRDGTVYVRESSDEVDEWRQINSIIAKPEVFGRKDDKEKIFEFLLTHARDSD 553
Query: 178 ELSVYSIVGHGGYGKTTLAQ 197
LSVY IVG GG GKTTL Q
Sbjct: 554 FLSVYPIVGLGGIGKTTLVQ 613
>TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type putative
resistance protein {Glycine max}, partial (61%)
Length = 676
Score = 163 bits (412), Expect = 3e-40
Identities = 94/226 (41%), Positives = 135/226 (59%), Gaps = 2/226 (0%)
Frame = +2
Query: 178 ELSVYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGK 237
++S SIVG GG GKTTLAQ V+ND R++ F+LK WV VS + + + +I+ G
Sbjct: 2 QVSTISIVGLGGMGKTTLAQLVYNDRRIQETFELKAWVYVSEYFDVIGLTNAILRK-FGT 178
Query: 238 NPHLSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRL 297
+ L+ +Q ++Q+ L YLLV+DDVW ++E W KL G G+ I++TTR
Sbjct: 179 AENSEDLDLLQWQLQKKLTGKNYLLVMDDVWKLNEESWEKLLLPFNYGSSGSKIILTTRD 358
Query: 298 DIVASIMGTSDAHHLASLSDDDIWSLFKQQAF-GENREERAELVAIGKKLVRKCVGSPLA 356
VA I+ +++ L L + D WSLFK+ AF G N E +L +IGK +V KC G PL
Sbjct: 359 KKVALIVKSTELVDLEQLKNKDCWSLFKRLAFHGSNVSEYPKLESIGKNIVDKCGGLPLT 538
Query: 357 AKVLGSSLCCTSNEHQWISVLESEFWNLPEVDS-IMSALRISYFNL 401
K +G+ L + +W +LE++ W L + DS I SALR+SY NL
Sbjct: 539 GKTMGNLLRKKFTQSEWEKILEADMWRLTDDDSNINSALRLSYHNL 676
>TC87724
Length = 720
Score = 160 bits (404), Expect = 3e-39
Identities = 95/194 (48%), Positives = 120/194 (60%), Gaps = 3/194 (1%)
Frame = +2
Query: 5 LLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKEWLQQ 64
LLG++ +NL S +Q E +T G+ Q LS L I+AVL+DAEKKQ ++K WLQ
Sbjct: 149 LLGVVFENLTSLLQNEFSTISGIKSKAQKLSDNLVRIKAVLEDAEKKQFKELSIKLWLQD 328
Query: 65 LRDAAYVLDDILDECSI-TLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDIAEE 123
L+DA YVLDDILDE SI + + G T F P I+ R IG R+KEI + +DDIAE
Sbjct: 329 LKDAVYVLDDILDEYSIKSCRLRG----CTSFKPKNIMFRHEIGNRLKEITRRLDDIAES 496
Query: 124 RMKFGLHVGVIERQPEDE--GRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELSV 181
+ KF L +G R+ D+ RQT S+I E KV+GR+ DKE I EFLL A DS+ LSV
Sbjct: 497 KNKFSLQMGGTLREIPDQVAEGRQTGSIIAEPKVFGREVDKEKIAEFLLTQARDSDFLSV 676
Query: 182 YSIVGHGGYGKTTL 195
Y IV K TL
Sbjct: 677 YPIVWFXWCWKNTL 718
>BF641864
Length = 656
Score = 156 bits (394), Expect = 4e-38
Identities = 91/188 (48%), Positives = 113/188 (59%), Gaps = 2/188 (1%)
Frame = +1
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MADALLG++ +NL S +Q E AT G+ + LS L I+AVL+DAEKKQ ++K+
Sbjct: 100 MADALLGVVSENLTSLLQNEFATISGIRSKARKLSDNLVHIKAVLEDAEKKQFKELSIKQ 279
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ L+DA YVL DILDE SI G + F PM I R IG R KEI + +DDI
Sbjct: 280 WLQDLKDAVYVLGDILDEYSI---ESGRLRGFNSFKPMNIAFRHEIGSRFKEITRRLDDI 450
Query: 121 AEERMKFGLHVGVIERQPEDE--GRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEE 178
AE + KF L +G R+ + RQT S ESK GRD DK+ FLL HA DS+
Sbjct: 451 AESKNKFSLQMGGTLREIPXQVAEGRQTXSTPLESKALGRDXDKKKXXXFLLTHAKDSDF 630
Query: 179 LSVYSIVG 186
+SVY I G
Sbjct: 631 ISVYPIXG 654
>TC81424
Length = 751
Score = 149 bits (375), Expect = 6e-36
Identities = 70/84 (83%), Positives = 78/84 (92%)
Frame = +1
Query: 786 EGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTIS 845
+GIPSLQKL+LFNFPS+KSLPD LGAMTSLQVL I +FP++ SLPDNFQQLQNLQ+L I
Sbjct: 1 KGIPSLQKLKLFNFPSLKSLPDCLGAMTSLQVLDIYNFPKVKSLPDNFQQLQNLQSLRIC 180
Query: 846 GCPILEKRCKRGIGEDWHKIAHIP 869
GCP+LEKRCKRGIGEDWHKIAHIP
Sbjct: 181 GCPMLEKRCKRGIGEDWHKIAHIP 252
Score = 38.9 bits (89), Expect = 0.009
Identities = 21/62 (33%), Positives = 34/62 (53%), Gaps = 1/62 (1%)
Frame = +1
Query: 717 KGLSSLRNMSVFSCSGFKSLSDGMRHLTCLETLHIYYCPQL-VFPHNMNSLASLRQLLLV 775
KG+ SL+ + +F+ KSL D + +T L+ L IY P++ P N L +L+ L +
Sbjct: 1 KGIPSLQKLKLFNFPSLKSLPDCLGAMTSLQVLDIYNFPKVKSLPDNFQQLQNLQSLRIC 180
Query: 776 EC 777
C
Sbjct: 181 GC 186
Score = 31.2 bits (69), Expect = 1.8
Identities = 24/86 (27%), Positives = 36/86 (40%)
Frame = +1
Query: 672 NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCS 731
+L+ L + F +LK LP LG +T+L+ L I ++
Sbjct: 13 SLQKLKLFNFPSLKSLPDCLGAMTSLQVLDI-------------------------YNFP 117
Query: 732 GFKSLSDGMRHLTCLETLHIYYCPQL 757
KSL D + L L++L I CP L
Sbjct: 118 KVKSLPDNFQQLQNLQSLRICGCPML 195
>BF647098 weakly similar to GP|22324956|gb| putative disease resistant
protein {Oryza sativa (japonica cultivar-group)},
partial (2%)
Length = 683
Score = 144 bits (363), Expect = 1e-34
Identities = 86/173 (49%), Positives = 106/173 (60%), Gaps = 3/173 (1%)
Frame = +2
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA ALLG++ +NL S +Q E +T G+ Q LS L I+AVL+DAEKKQ ++K
Sbjct: 173 MACALLGVVFENLTSLLQNEFSTISGIKSKAQKLSDNLVHIKAVLEDAEKKQFKELSIKL 352
Query: 61 WLQQLRDAAYVLDDILDECSI-TLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDD 119
WLQ L+DA YVLDDILDE SI + + G T F P I+ R IG R+KEI + +DD
Sbjct: 353 WLQDLKDAVYVLDDILDEYSIESCRLRG----FTSFKPKNIMFRHEIGNRLKEITRRLDD 520
Query: 120 IAEERMKFGLHVGVIERQPEDE--GRRQTTSVITESKVYGRDKDKEHIVEFLL 170
IAE + KF L G R D+ RQT+S ESK GRD D E IVEFLL
Sbjct: 521 IAERKNKFSLQTGETLRVIPDQVAEGRQTSSTPXESKALGRDDDXEKIVEFLL 679
>TC91142 weakly similar to PIR|T02213|T02213 NBS-LRR type resistance protein
- rice (fragment), partial (6%)
Length = 944
Score = 143 bits (361), Expect = 3e-34
Identities = 85/133 (63%), Positives = 92/133 (68%), Gaps = 18/133 (13%)
Frame = +1
Query: 403 LSLRPCFAFCAVFPKGFEMVKENLIHLWMAN-----------GLVTSRGNLQMEHVGD-- 449
L L C + FPK F +K+ L HL + N G +TS L + VG
Sbjct: 550 LKLERCHLLSS-FPKQFTKLKD-LRHLMIKNCHSLISAPFRIGQLTSLKTLTIFIVGSKT 723
Query: 450 -----ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVD 504
+LHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVD
Sbjct: 724 GYGLAQLHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVD 903
Query: 505 AERVLEALEPHSS 517
AERVLEALEPHSS
Sbjct: 904 AERVLEALEPHSS 942
Score = 56.6 bits (135), Expect = 4e-08
Identities = 25/30 (83%), Positives = 27/30 (89%)
Frame = +1
Query: 424 ENLIHLWMANGLVTSRGNLQMEHVGDELHN 453
ENLIHLWMANG VTS GNLQMEHVGD++ N
Sbjct: 1 ENLIHLWMANGRVTSTGNLQMEHVGDDVWN 90
>TC78027 similar to PIR|D86327|D86327 protein F18O14.17 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 2226
Score = 104 bits (259), Expect(2) = 3e-34
Identities = 75/185 (40%), Positives = 99/185 (52%), Gaps = 4/185 (2%)
Frame = +1
Query: 507 RVLEALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLP 566
+ LE L+PHS+ LK +N Y G PSW+ IL L+S+ L C LP GKLP
Sbjct: 34 KYLEVLQPHSN-LKCLTINYYEGLSLPSWI---IILSNLISLELEICNKIVRLPLLGKLP 201
Query: 567 CLTILYLSGMRYIKYIDDDLYEPETE-KAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLL 625
L L L GM +KY+DDD E E F SL++L+L LPN+E +L+V+ EM P L
Sbjct: 202 SLKKLRLYGMNNLKYLDDDESEYGMEVSVFPSLEELNLKSLPNIEGLLKVERGEMFPCLS 381
Query: 626 NLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDV---AGNNLKSLSISKFA 682
LDI + P+L L L S++SL N ELL+S + +G + SL F
Sbjct: 382 KLDIWDCPELGLPCLPSLKSLHLWECNNELLRSISTFRGLTQLTLNSGEGITSLPEEMFK 561
Query: 683 NLKEL 687
NL L
Sbjct: 562 NLTSL 576
Score = 60.8 bits (146), Expect(2) = 7e-18
Identities = 33/70 (47%), Positives = 43/70 (61%), Gaps = 2/70 (2%)
Frame = +3
Query: 802 IKSLPD--WLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIG 859
++SLP+ W G + SL+ L I L LP+ + L +L+ L I CP LE+RCK G
Sbjct: 606 LESLPEQNWEG-LQSLRALQIWGCRGLRCLPEGIRHLTSLELLDIIDCPTLEERCKEGTW 782
Query: 860 EDWHKIAHIP 869
EDW KIAHIP
Sbjct: 783 EDWDKIAHIP 812
Score = 60.1 bits (144), Expect(2) = 3e-34
Identities = 29/59 (49%), Positives = 37/59 (62%)
Frame = +3
Query: 699 SLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLTCLETLHIYYCPQL 757
SL I CNE+ES E +GL SLR + ++ C G + L +G+RHLT LE L I CP L
Sbjct: 579 SLCINCCNELESLPEQNWEGLQSLRALQIWGCRGLRCLPEGIRHLTSLELLDIIDCPTL 755
Score = 48.5 bits (114), Expect(2) = 7e-18
Identities = 50/170 (29%), Positives = 71/170 (41%), Gaps = 33/170 (19%)
Frame = +1
Query: 671 NNLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSC 730
+NLK L+I+ + L LP + L+ L SL +E CN++ LL L SL+ + ++
Sbjct: 61 SNLKCLTINYYEGLS-LPSWIIILSNLISLELEICNKIVRLP--LLGKLPSLKKLRLYGM 231
Query: 731 SGFKSLSD-----GMR----------------------------HLTCLETLHIYYCPQL 757
+ K L D GM CL L I+ CP+L
Sbjct: 232 NNLKYLDDDESEYGMEVSVFPSLEELNLKSLPNIEGLLKVERGEMFPCLSKLDIWDCPEL 411
Query: 758 VFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKLRLFNFPSIKSLPD 807
P L SL+ L L ECN +L I L +L L + I SLP+
Sbjct: 412 GLP----CLPSLKSLHLWECNNELLRSISTFRGLTQLTLNSGEGITSLPE 549
>AW586179
Length = 590
Score = 143 bits (360), Expect = 3e-34
Identities = 87/180 (48%), Positives = 111/180 (61%), Gaps = 4/180 (2%)
Frame = +1
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
M DALLG++ +NL + + +E +T G+ Q LS L I+AVL+DAEKK ++K
Sbjct: 64 MDDALLGLVYENLTAILHKEYSTISGINLKVQKLSNNLVHIKAVLEDAEKKHFKELSIKL 243
Query: 61 WLQQLRDAAYVLDDILDECSI-TLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDD 119
WLQ L+D YVLDDILDE SI + + G T F P I+ R IG KEI + +DD
Sbjct: 244 WLQDLKDGVYVLDDILDEYSIKSCRLRG----FTSFKPKNIMFRHEIGNTFKEITRRLDD 411
Query: 120 IAEERMKFGLHVGVIERQ-PED--EGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDS 176
IAE + KF L +G R+ P EG RQT S+I E KV+GR+ DKE IVEFLL A DS
Sbjct: 412 IAESKNKFSLQMGGTLREIPHQVAEG-RQTGSIIAEPKVFGREVDKEKIVEFLLTQARDS 588
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,088,337
Number of Sequences: 36976
Number of extensions: 438862
Number of successful extensions: 2717
Number of sequences better than 10.0: 252
Number of HSP's better than 10.0 without gapping: 2435
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2579
length of query: 887
length of database: 9,014,727
effective HSP length: 105
effective length of query: 782
effective length of database: 5,132,247
effective search space: 4013417154
effective search space used: 4013417154
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146556.4


