

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146555.15 - phase: 0 /pseudo
(552 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
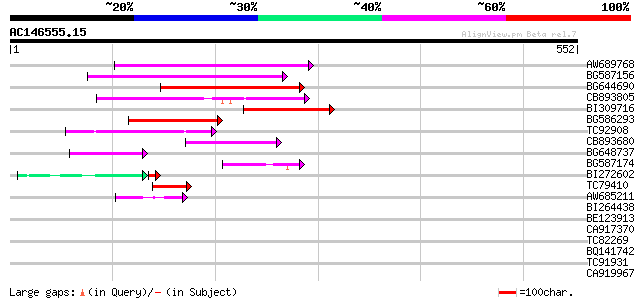
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 170 1e-42
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 140 1e-33
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 116 2e-26
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 109 2e-24
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 82 4e-16
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 82 5e-16
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 76 3e-14
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 62 6e-10
BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberos... 55 9e-08
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 48 9e-06
BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%) 42 2e-04
TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-l... 42 5e-04
AW685211 weakly similar to GP|4539662|gb| polyprotein {Sorghum b... 42 8e-04
BI264438 similar to GP|21427467|gb actin-related protein 6 {Arab... 36 0.044
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 34 0.13
CA917370 similar to GP|15010620|gb AT4g08960/T3H13_2 {Arabidopsi... 33 0.28
TC82269 weakly similar to GP|14581464|gb|AAK31804.1 protocadheri... 33 0.37
BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvo... 32 0.63
TC91931 weakly similar to PIR|T47879|T47879 hypothetical protein... 32 0.63
CA919967 similar to GP|21522866|em cDNA coding for hydroxyphenyl... 32 0.63
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 170 bits (430), Expect = 1e-42
Identities = 88/193 (45%), Positives = 116/193 (59%)
Frame = +1
Query: 103 WKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQ 162
W Q M+ E L TW +V LPP K IGC WV +VK N DGS+ ++K LVAKG++Q
Sbjct: 97 WLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQ 276
Query: 163 IEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVS 222
G DY +TFS V K TIRL++ +A + Q+D+NNAFL+G L E+VYM+ P G
Sbjct: 277 TLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFE 456
Query: 223 TSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLV 282
+ + VCKL+KSLYGLK A R WYE LT I G+ ++ + SL + I L +
Sbjct: 457 AANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXI 636
Query: 283 YVYDITLAGDFLS 295
YV DI + G S
Sbjct: 637 YVDDILITGSSAS 675
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 140 bits (354), Expect = 1e-33
Identities = 76/196 (38%), Positives = 117/196 (58%), Gaps = 1/196 (0%)
Frame = -1
Query: 76 KSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCI 135
++H F ++L P+SY+EA++ WK+ + E + + TW +LP K +
Sbjct: 603 EAHCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSR 424
Query: 136 WVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FL 195
W+ +K+ DGSIER K LVA+G+ G DY +TF+ VAK+ TIR+V++LA L
Sbjct: 423 WIFTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGL 244
Query: 196 HQLDVNNAFLHGDLHEDVYMAIPPGVS-TSKPNQVCKLSKSLYGLKPASRKWYEKLTCLP 254
Q+DV NAFL G+L ++VYM PPG+ K V +L K++YGLK + R WY KL+
Sbjct: 243 WQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTL 64
Query: 255 ITNGYQQATSNASLFT 270
G++++ + +LFT
Sbjct: 63 NGRGFRKSELDHTLFT 16
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 116 bits (291), Expect = 2e-26
Identities = 62/141 (43%), Positives = 94/141 (65%), Gaps = 1/141 (0%)
Frame = -2
Query: 148 IERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHG 207
I R K LV +GYNQ EG+DY + FS VA++ IR++IA A+ L+Q+DV +AF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 208 DLHEDVYMAIPPGVSTSK-PNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNA 266
DL E+V++ PPG ++ PN V +L+K+LYGLK A R WYE+L+ + NG+++ +
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 267 SLFTKKNLDSFIMLLVYVYDI 287
+LF K +++ VYV DI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 109 bits (273), Expect = 2e-24
Identities = 68/217 (31%), Positives = 117/217 (53%), Gaps = 9/217 (4%)
Frame = +3
Query: 85 LVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNV 144
L ++P +++EA+K + W+ M NE+ ++ TW++ DL +K IG W+ K K N
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 145 DGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAF 204
+G IE+YK LVAKGY+Q G+DY + F+ VA+ TIR+VIALA+ Q+ +
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA-------QIKRDGVC 362
Query: 205 L------HGDLHEDV--YMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPIT 256
+ H + + ++ I V + ++ ++LYGLK A R WY ++
Sbjct: 363 IS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS-*RVKRALYGLKQAPRAWYSRIEAYFTK 539
Query: 257 NGYQQATSNASLFTKKNLDSFIMLL-VYVYDITLAGD 292
G+++ +LF K + I+++ +YV D+ G+
Sbjct: 540 EGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGN 650
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 82.4 bits (202), Expect = 4e-16
Identities = 48/89 (53%), Positives = 60/89 (66%)
Frame = +2
Query: 228 QVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDI 287
+VC+L KS+YGLK ASR+WY KL+ I+ GY Q++S+ SLFTK SF LLVYV DI
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 288 TLAGDFLSEITFIKNALNQASKSKILVSL 316
LAG+ +SEI +K L K K L SL
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSL 283
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 82.0 bits (201), Expect = 5e-16
Identities = 41/92 (44%), Positives = 61/92 (65%)
Frame = +2
Query: 116 QTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLV 175
Q T K+V P KPIG W+ K+K N DG++ +YK LVAKGY + +G+D+ + F+ V
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 176 AKVTTIRLVIALASINY*FLHQLDVNNAFLHG 207
++ TI L++ALA+ N +H +DV AFL+G
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 76.3 bits (186), Expect = 3e-14
Identities = 47/147 (31%), Positives = 79/147 (52%)
Frame = +2
Query: 55 TTNNSHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTL 114
T N+S Y I +++ +++ +K + A ++ S EPK+Y +A + W M+ E+ L
Sbjct: 176 TLNSSSKSYHIFHYVGYSFSAKHRASLA-AITSNIEPKNYVQAAQ*QEWLAAMEQEIQVL 352
Query: 115 DQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSL 174
++ T + L K + C V K+ H +G IE+YK LVAK + Q+EG D+ D L
Sbjct: 353 EENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGEDF*D-LCL 529
Query: 175 VAKVTTIRLVIALASINY*FLHQLDVN 201
K R ++ +A+ * LH +DV+
Sbjct: 530 SNKDDNCRCLLTIAAAKG*QLHLMDVS 610
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 62.0 bits (149), Expect = 6e-10
Identities = 36/93 (38%), Positives = 49/93 (51%)
Frame = -2
Query: 172 FSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCK 231
F + K+ TI ++++ +I +L LDV AFL GDL ED+YM P G S V K
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGK 375
Query: 232 LSKSLYGLKPASRKWYEKLTCLPITNGYQQATS 264
L KS+YGLK R+ L L G++ S
Sbjct: 374 LKKSMYGLKQGPRQCI*SLKALCTRKGFRSEIS 276
>BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberosum}, partial
(10%)
Length = 804
Score = 54.7 bits (130), Expect = 9e-08
Identities = 25/76 (32%), Positives = 40/76 (51%)
Frame = +3
Query: 59 SHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTG 118
S +PI F+S+ LS H F +L P S E++ + WK+ + E+ L++
Sbjct: 573 SCTSHPICKFISYDKLSPHHLAFVSNLDKIQIPNSVHESLTNP*WKKAILGEIHALEKNE 752
Query: 119 TWKIVDLPPSSKPIGC 134
TW++ DLP P+GC
Sbjct: 753 TWELSDLPSGKHPMGC 800
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 48.1 bits (113), Expect = 9e-06
Identities = 28/88 (31%), Positives = 47/88 (52%), Gaps = 8/88 (9%)
Frame = -1
Query: 208 DLHEDVYMAIPPG-VSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNA 266
+L E +YM P G + K + VCKL KSLYGLK + R+WY++ + Y+ + +
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRF------DSYRSSWATT 88
Query: 267 SLF-------TKKNLDSFIMLLVYVYDI 287
+ T+ + +I L++YV D+
Sbjct: 87 GVLMTVVST*TR*RMSRYIYLVLYVDDM 4
>BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%)
Length = 488
Score = 42.0 bits (97), Expect(2) = 2e-04
Identities = 31/127 (24%), Positives = 51/127 (39%)
Frame = +1
Query: 8 PTTSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTTNNSHVQYPISN 67
PT++ IT TSP+PP PPP N T N+ T + +++ I
Sbjct: 85 PTSNELANIT-TSPTPPPPPPPTARPHNNHTT---------NVGGIITRSQNNIVKTIKK 234
Query: 68 FLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPP 127
H + + EP + +A++ W+ +MQ+E L TW +V
Sbjct: 235 LNLH-----------VRPLCPIEPSNITQALRDHDWRSIMQDEFDALQNNNTWDLVSRSF 381
Query: 128 SSKPIGC 134
+ +GC
Sbjct: 382 AQNLVGC 402
Score = 21.2 bits (43), Expect(2) = 2e-04
Identities = 7/12 (58%), Positives = 9/12 (74%)
Frame = +2
Query: 136 WVSKVKHNVDGS 147
WV ++K N DGS
Sbjct: 407 WVFRIKRNPDGS 442
>TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-like protein
[imported] - Arabidopsis thaliana, partial (84%)
Length = 1002
Score = 42.4 bits (98), Expect = 5e-04
Identities = 21/38 (55%), Positives = 25/38 (65%)
Frame = +3
Query: 140 VKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAK 177
+K N DGS Y LVAKG++Q G+DY D FSLV K
Sbjct: 768 LKRNSDGSTLYYTTRLVAKGFHQRSGIDYKDQFSLVVK 881
>AW685211 weakly similar to GP|4539662|gb| polyprotein {Sorghum bicolor},
partial (2%)
Length = 388
Score = 41.6 bits (96), Expect = 8e-04
Identities = 26/70 (37%), Positives = 34/70 (48%)
Frame = -2
Query: 104 KQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQI 163
+Q M E+T L+ TW I+ LP KH R KV + AKGY+++
Sbjct: 225 QQAMIGEMTALESNRTWTIIPLPLEMP---------YKH-------RLKVRMAAKGYSKV 94
Query: 164 EGLDYFDTFS 173
GL Y DTFS
Sbjct: 93 YGLHYNDTFS 64
>BI264438 similar to GP|21427467|gb actin-related protein 6 {Arabidopsis
thaliana}, partial (43%)
Length = 695
Score = 35.8 bits (81), Expect = 0.044
Identities = 34/92 (36%), Positives = 41/92 (43%), Gaps = 9/92 (9%)
Frame = +1
Query: 2 TAHTHIPTTSPSPPITKTSPSPPFIPPPIRIT----TRNKI-----TPTYM*DYICNIPT 52
T T +P P PP KTSP PP P I T T N I +P + N P
Sbjct: 13 TLSTPLPPPPPPPPPNKTSPPPPSAAPSIAAT*STPTSNAISGPTFSPLSSASTLLN-PL 189
Query: 53 TSTTNNSHVQYPISNFLSHTYLSKSHSIFAMS 84
+S+ N S + P N LS T S SIF +S
Sbjct: 190 SSSLNPSSLSLPF-NTLS-TNSSSKISIFPLS 279
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 34.3 bits (77), Expect = 0.13
Identities = 24/74 (32%), Positives = 39/74 (52%), Gaps = 1/74 (1%)
Frame = +1
Query: 244 RKWYEKLTCLPITNGYQQATSNASLFTKKNLD-SFIMLLVYVYDITLAGDFLSEITFIKN 302
R W+++ T + GY Q ++ ++F K + +L+VYV DI L GD I +KN
Sbjct: 10 RDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKRLKN 189
Query: 303 ALNQASKSKILVSL 316
L + + K L +L
Sbjct: 190 LLAEEFEIKDLGNL 231
>CA917370 similar to GP|15010620|gb AT4g08960/T3H13_2 {Arabidopsis
thaliana}, partial (26%)
Length = 604
Score = 33.1 bits (74), Expect = 0.28
Identities = 22/65 (33%), Positives = 30/65 (45%), Gaps = 11/65 (16%)
Frame = +1
Query: 1 FTAHTHIPTTSPSPPITKTSPSPPFIPPPIRITT-----------RNKITPTYM*DYICN 49
F+ HT P T+ IT T P P IPP ++TT N T T + + N
Sbjct: 34 FSHHTASPKTNHVRRITPTPPPPRRIPPHSKLTTHHRHLLQVRRPNNLRTTTTLPQLLRN 213
Query: 50 IPTTS 54
+PTT+
Sbjct: 214LPTTN 228
>TC82269 weakly similar to GP|14581464|gb|AAK31804.1 protocadherin 15 {Homo
sapiens}, partial (1%)
Length = 728
Score = 32.7 bits (73), Expect = 0.37
Identities = 22/74 (29%), Positives = 36/74 (47%), Gaps = 5/74 (6%)
Frame = +2
Query: 9 TTSPSPPITKTSPSPPFI----PPPIRITTRNKI-TPTYM*DYICNIPTTSTTNNSHVQY 63
+TSP P I+ PSPP PPP ++ T N + +P + + N + TT+ H
Sbjct: 179 STSPIPQISHQPPSPPPTPPNPPPPSKMKTLNPVHSPKWSNNSSTNATISQTTDTHHKST 358
Query: 64 PISNFLSHTYLSKS 77
S + T L+++
Sbjct: 359 KSSTKTTFTKLNQT 400
>BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvox carteri,
partial (9%)
Length = 1337
Score = 32.0 bits (71), Expect = 0.63
Identities = 14/26 (53%), Positives = 16/26 (60%), Gaps = 4/26 (15%)
Frame = -3
Query: 8 PTTSPSPPITKTSPS----PPFIPPP 29
PT P+PP+T PS PPF PPP
Sbjct: 96 PTPKPNPPLTPPPPSPPPHPPFPPPP 19
>TC91931 weakly similar to PIR|T47879|T47879 hypothetical protein T4C21.90 -
Arabidopsis thaliana, partial (10%)
Length = 714
Score = 32.0 bits (71), Expect = 0.63
Identities = 29/78 (37%), Positives = 37/78 (47%), Gaps = 2/78 (2%)
Frame = +1
Query: 7 IPTTSPSPPITKTSPSPPFIPPPIRITTRNKIT--PTYM*DYICNIPTTSTTNNSHVQYP 64
+P T+PS P T S SPP P T N++T T D I + P T T + Q P
Sbjct: 313 LPETTPSSPKTTPSSSPPKTTPSF---TPNQLTLPETASFDLIDSSPKT-TPLFTQTQSP 480
Query: 65 ISNFLSHTYLSKSHSIFA 82
+ SH +S SIFA
Sbjct: 481 LPKTTSHDLIS---SIFA 525
>CA919967 similar to GP|21522866|em cDNA coding for hydroxyphenylpyruvate
dioxygenase {Arabidopsis thaliana}, partial (40%)
Length = 778
Score = 32.0 bits (71), Expect = 0.63
Identities = 24/91 (26%), Positives = 38/91 (41%), Gaps = 9/91 (9%)
Frame = +2
Query: 6 HIPTTSPSPPITKTSPSPPFIPP---PIRITTRNKITPTYM*DYICNIPTTSTTNNSHVQ 62
HIP SP + + P PP +PP P + T P+ + N P+ S +H
Sbjct: 497 HIPLPFLSPRLPQPPPFPPSLPPLASPSALHTALPFAPSLLKSTTQNSPSLSA*TTAHFH 676
Query: 63 YPISNF------LSHTYLSKSHSIFAMSLVS 87
+ + +F L S S S +A S+ +
Sbjct: 677 HLLPSFSKTASNLLKVISSASTSFYATSVTT 769
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.336 0.146 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,006,980
Number of Sequences: 36976
Number of extensions: 305204
Number of successful extensions: 6747
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 4621
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6241
length of query: 552
length of database: 9,014,727
effective HSP length: 101
effective length of query: 451
effective length of database: 5,280,151
effective search space: 2381348101
effective search space used: 2381348101
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146555.15


