

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146554.10 + phase: 2 /partial
(83 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
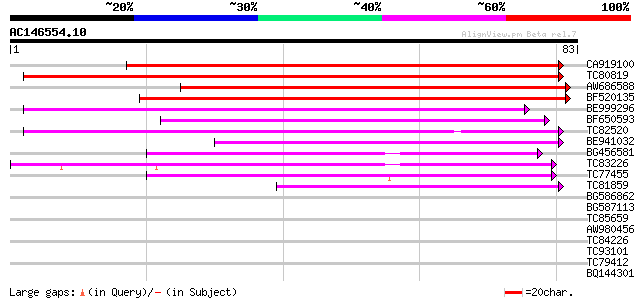
Score E
Sequences producing significant alignments: (bits) Value
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 96 3e-21
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 80 2e-16
AW686588 76 2e-15
BF520135 64 8e-12
BE999296 58 6e-10
BF650593 56 2e-09
TC82520 54 1e-08
BE941032 53 2e-08
BG456581 42 4e-05
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 42 4e-05
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 41 7e-05
TC81859 similar to GP|18491135|gb|AAL69536.1 At1g04290/F19P19_27... 39 5e-04
BG586862 37 0.002
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 31 0.075
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 29 0.28
AW980456 28 0.63
TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-... 28 0.82
TC93101 27 1.1
TC79412 similar to GP|15809663|gb|AAL07275.1 COP9 complex subuni... 27 1.8
BQ144301 similar to GP|15277263|db alternative name: G2~unknown ... 26 3.1
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 95.5 bits (236), Expect = 3e-21
Identities = 42/64 (65%), Positives = 53/64 (82%)
Frame = -2
Query: 18 SELIWHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVS 77
+E+IWHR VPLKVS+ AW LLR+RLPTK+NL+ RG + +A LCVSGC A+E+AQHLF+S
Sbjct: 593 AEMIWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGLCVSGCGALESAQHLFLS 414
Query: 78 CPIF 81
C F
Sbjct: 413 CSYF 402
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 79.7 bits (195), Expect = 2e-16
Identities = 36/79 (45%), Positives = 50/79 (62%)
Frame = +2
Query: 3 YDLLTADDNNTDVAVSELIWHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCV 62
Y +T+ + +D ++ + +WH+H+P KVS+ W LLRNRLPTK NLV RG L CV
Sbjct: 572 YRYITSTGHISDRSLVDDVWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVLLATNAACV 751
Query: 63 SGCWAVETAQHLFVSCPIF 81
GC E+ HLF+ C +F
Sbjct: 752 CGCVDSESTTHLFLHCNVF 808
>AW686588
Length = 567
Score = 76.3 bits (186), Expect = 2e-15
Identities = 38/57 (66%), Positives = 42/57 (73%)
Frame = +1
Query: 26 VPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSCPIFG 82
VPLKVSILAW L+R+RLPTKANLV R LA +A CV GC ETA HLF+ C FG
Sbjct: 142 VPLKVSILAWRLIRDRLPTKANLVRRRCLAVEAAGCVVGCGIAETANHLFLHCATFG 312
>BF520135
Length = 202
Score = 64.3 bits (155), Expect = 8e-12
Identities = 29/63 (46%), Positives = 39/63 (61%)
Frame = +3
Query: 20 LIWHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSCP 79
L+W + KVSI AW L RLPTKAN+ RG + DA +CV+ C +E+ HLF+ C
Sbjct: 12 LLWRKEDLSKVSIFAWRLFHGRLPTKANVFKRGIVHHDAHMCVTRCRLIESDVHLFLHCD 191
Query: 80 IFG 82
+ G
Sbjct: 192VLG 200
>BE999296
Length = 384
Score = 58.2 bits (139), Expect = 6e-10
Identities = 29/74 (39%), Positives = 38/74 (51%)
Frame = -1
Query: 3 YDLLTADDNNTDVAVSELIWHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCV 62
Y L + D D + +WH H+PLKV +LRN LPTK N V R + + LC
Sbjct: 294 YQFLMSADAPLDREYIDSVWHNHIPLKVCFFVLRVLRNCLPTKDNFVRRRVIHEEHMLCP 115
Query: 63 SGCWAVETAQHLFV 76
+GC ET LF+
Sbjct: 114 TGCSFKETTDDLFL 73
>BF650593
Length = 486
Score = 56.2 bits (134), Expect = 2e-09
Identities = 26/57 (45%), Positives = 32/57 (55%)
Frame = +3
Query: 23 HRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSCP 79
H+ VPLKVS L W L +N L T+ NL RG L ++ CV C E+ H F CP
Sbjct: 297 HKSVPLKVSCLVWRLFQNXLATRDNLSKRGVLDQNSIXCVXDCGREESVSHFFFECP 467
>TC82520
Length = 833
Score = 53.9 bits (128), Expect = 1e-08
Identities = 29/79 (36%), Positives = 39/79 (48%)
Frame = +3
Query: 3 YDLLTADDNNTDVAVSELIWHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCV 62
Y LT D + +W +++P KVS+ W L NRLPTK NL+ R L C+
Sbjct: 132 YRFLTISGQPLDRNQVDDVWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHVLQQTHTACI 311
Query: 63 SGCWAVETAQHLFVSCPIF 81
SG A+ QH+ IF
Sbjct: 312 SGV-AIRKRQHICFYIVIF 365
Score = 29.3 bits (64), Expect = 0.28
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +2
Query: 64 GCWAVETAQHLFVSCPIFG 82
GC ETA HLF+ C IFG
Sbjct: 314 GCGDSETATHLFLHCDIFG 370
>BE941032
Length = 435
Score = 52.8 bits (125), Expect = 2e-08
Identities = 24/51 (47%), Positives = 30/51 (58%)
Frame = +2
Query: 31 SILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSCPIF 81
SI WC+L NR PTK NL+ RG ++ Q CV C + A HLF+ C F
Sbjct: 146 SIFLWCVLLNRFPTKDNLLKRGVISAIYQSCVGECGNLYDATHLFLHCNFF 298
>BG456581
Length = 683
Score = 42.0 bits (97), Expect = 4e-05
Identities = 22/58 (37%), Positives = 30/58 (50%)
Frame = +2
Query: 21 IWHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSC 78
IW+ +P S + W L +RLPT NL +RG +C VET+ HLF+ C
Sbjct: 80 IWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCAL--VSMCSFCLEQVETSDHLFLRC 247
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 42.0 bits (97), Expect = 4e-05
Identities = 29/89 (32%), Positives = 41/89 (45%), Gaps = 9/89 (10%)
Frame = +3
Query: 1 TGYDLL----TADDNNTDVAVSEL-----IWHRHVPLKVSILAWCLLRNRLPTKANLVAR 51
+GY+ L T NNT + E IW H + +L W +L + LP +++L R
Sbjct: 42 SGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRILNDSLPVRSSLRKR 221
Query: 52 GTLAPDAQLCVSGCWAVETAQHLFVSCPI 80
G LC ET HLF+SCP+
Sbjct: 222 GIQC--YPLCPRCHSKTETITHLFMSCPL 302
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 41.2 bits (95), Expect = 7e-05
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Frame = -2
Query: 21 IWHRHVPLKVSILAWCLLRNRLPTKANLVARGTL-APDAQLCVSGCWAVETAQHLFVSCP 79
+W P KV +W L +R+PT NL R L D++ CV ET HLF+ C
Sbjct: 322 LWKSKAPAKVLAFSWTLFLDRIPTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHCD 143
Query: 80 I 80
+
Sbjct: 142 V 140
>TC81859 similar to GP|18491135|gb|AAL69536.1 At1g04290/F19P19_27
{Arabidopsis thaliana}, partial (65%)
Length = 960
Score = 38.5 bits (88), Expect = 5e-04
Identities = 19/42 (45%), Positives = 25/42 (59%)
Frame = +2
Query: 40 NRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSCPIF 81
+RLPTK N++ R D+ CV G ET+Q LF CP+F
Sbjct: 617 DRLPTKYNVLPREINNLDSSFCVGGWEVNETSQSLFF*CPLF 742
>BG586862
Length = 804
Score = 36.6 bits (83), Expect = 0.002
Identities = 24/65 (36%), Positives = 33/65 (49%), Gaps = 1/65 (1%)
Frame = -1
Query: 17 VSELIWH-RHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLF 75
+ E +W + +P S L W LL N LP K L RG + LC +ET QHLF
Sbjct: 666 IREKVWGIKTIPRHKSFL-WRLLHNALPVKDELHKRGIRC--SLLCPRCESKIETVQHLF 496
Query: 76 VSCPI 80
++C +
Sbjct: 495 LNCEV 481
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 31.2 bits (69), Expect = 0.075
Identities = 18/59 (30%), Positives = 27/59 (45%)
Frame = -3
Query: 21 IWHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSCP 79
+W + KV W + N LPT AN+ +R ++ D G + ET H+ CP
Sbjct: 288 VWKYNTSPKVRHFLWRCISNSLPTAANMRSR-HISKDGSCSRCGMES-ETVNHILFQCP 118
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 29.3 bits (64), Expect = 0.28
Identities = 18/43 (41%), Positives = 21/43 (47%)
Frame = +2
Query: 22 WHRHVPLKVSILAWCLLRNRLPTKANLVARGTLAPDAQLCVSG 64
W R V KV L C LRN L + V ++PD LC SG
Sbjct: 572 WDRTV--KVWNLTNCKLRNTLAGHSGYVNTVAVSPDGSLCASG 694
>AW980456
Length = 779
Score = 28.1 bits (61), Expect = 0.63
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = -2
Query: 21 IWHRHVPLKVSILAWCLLRNRLPTKANL 48
IW VP KV L W + R LPT+ L
Sbjct: 88 IWKLKVPPKVQNLVWRMCRGCLPTRIRL 5
>TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-like
protein GS0-1 {Glycine max}, partial (72%)
Length = 665
Score = 27.7 bits (60), Expect = 0.82
Identities = 10/29 (34%), Positives = 18/29 (61%)
Frame = -1
Query: 35 WCLLRNRLPTKANLVARGTLAPDAQLCVS 63
WCL+++ LP+K ++R T+ C+S
Sbjct: 527 WCLIKSYLPSKLVFISRRTMVARQVSCLS 441
>TC93101
Length = 675
Score = 27.3 bits (59), Expect = 1.1
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = -1
Query: 42 LPTKANLVARGTLAPDAQLCVSGCWAVETAQHLFVSC 78
LP + L RG P LC + +ET H+F+SC
Sbjct: 663 LPVRXELNKRGVNCPP--LCPRCYFNLETTNHIFMSC 559
>TC79412 similar to GP|15809663|gb|AAL07275.1 COP9 complex subunit 6
{Arabidopsis thaliana}, partial (93%)
Length = 1380
Score = 26.6 bits (57), Expect = 1.8
Identities = 15/31 (48%), Positives = 18/31 (57%)
Frame = +3
Query: 1 TGYDLLTADDNNTDVAVSELIWHRHVPLKVS 31
T + LLT T V V L++HRHV L VS
Sbjct: 213 TLHSLLTTTPPTTAVTV*FLLFHRHVSLVVS 305
>BQ144301 similar to GP|15277263|db alternative name: G2~unknown function
{Homo sapiens}, partial (1%)
Length = 1250
Score = 25.8 bits (55), Expect = 3.1
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = -1
Query: 34 AWCLLRNRLPTKANLVARGTLAP 56
AWC +RNR + N + R T P
Sbjct: 329 AWCQMRNRATSTGNKIWRNTNQP 261
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.137 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,123,669
Number of Sequences: 36976
Number of extensions: 40271
Number of successful extensions: 299
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 297
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 297
length of query: 83
length of database: 9,014,727
effective HSP length: 59
effective length of query: 24
effective length of database: 6,833,143
effective search space: 163995432
effective search space used: 163995432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC146554.10


