

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.5 - phase: 1 /pseudo
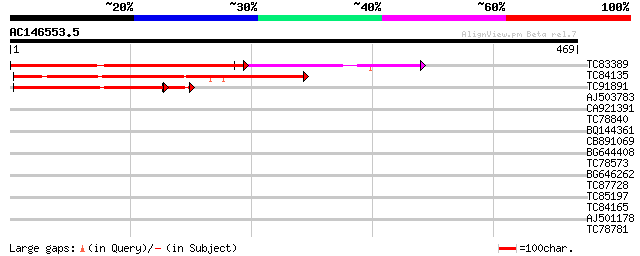
(469 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83389 weakly similar to GP|5231145|gb|AAD41092.1| xyloglucan f... 268 3e-72
TC84135 similar to SP|Q9M5Q1|FUT1_PEA Galactoside 2-L-fucosyltra... 204 6e-53
TC91891 weakly similar to GP|5231145|gb|AAD41092.1| xyloglucan f... 166 4e-43
AJ503783 similar to SP|O22307|C7DB_ Cytochrome P450 71D11 (EC 1.... 31 1.2
CA921391 similar to GP|1669591|dbj O-methyltransferase {Glycyrrh... 29 3.4
TC78840 similar to GP|21689605|gb|AAM67424.1 AT4g27690/T29A15_18... 29 4.4
BQ144361 28 5.8
CB891069 similar to PIR|E86428|E8 probable ABC transporter [impo... 28 5.8
BG644408 homologue to PIR|T07371|T07 dnaJ protein homolog - pota... 28 7.5
TC78573 similar to GP|20197309|gb|AAC63626.2 unknown protein {Ar... 28 7.5
BG646262 similar to PIR|T06036|T06 hypothetical protein T24A18.1... 28 9.9
TC87728 similar to GP|21554610|gb|AAM63632.1 unknown {Arabidopsi... 28 9.9
TC85197 GP|1435157|emb|CAA58445.1 histone H3 variant H3.3 {Lycop... 28 9.9
TC84165 similar to GP|21539497|gb|AAM53301.1 putative protein {A... 28 9.9
AJ501178 28 9.9
TC78781 weakly similar to PIR|T02291|T02291 hypothetical protein... 28 9.9
>TC83389 weakly similar to GP|5231145|gb|AAD41092.1| xyloglucan
fucosyltransferase {Arabidopsis thaliana}, partial (31%)
Length = 1065
Score = 268 bits (686), Expect = 3e-72
Identities = 128/198 (64%), Positives = 156/198 (78%), Gaps = 1/198 (0%)
Frame = +1
Query: 1 DEKEKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTR 60
++KEKLL+GLL SGF++ASCVSRLQSH Y K SPHKPS YLISKLR YEE+HRRCGPN +
Sbjct: 55 NDKEKLLNGLLASGFEDASCVSRLQSHLYRKVSPHKPSSYLISKLRNYEEIHRRCGPNGK 234
Query: 61 AYNEDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFG 120
Y++ M+KI S A+ CKY+IW PANGL NQ++SMA++F+YALLT RVMLV+F
Sbjct: 235 DYHKSMRKILNS-----GDASICKYLIWTPANGLXNQMVSMAATFMYALLTNRVMLVRFR 399
Query: 121 KDKEGLFCEPFLNSTWLLPEKSPFWNAEKVQTYQSTIKMGGANTLNE-DFPSALHVNLGY 179
KDK+G+FCEPFLNSTWLLPEKSPFWN V+TYQS I+ AN ++ D PS+L +NL Y
Sbjct: 400 KDKQGIFCEPFLNSTWLLPEKSPFWNENHVETYQSLIEKEKANNNSKLDLPSSLFLNLQY 579
Query: 180 SPTSEERFFHCDHNQFLL 197
P E+FFHCDH Q L+
Sbjct: 580 RPNLPEKFFHCDHRQALI 633
Score = 117 bits (294), Expect = 7e-27
Identities = 73/165 (44%), Positives = 92/165 (55%), Gaps = 7/165 (4%)
Frame = +2
Query: 187 FFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITSTFHHLGRYLFHP 246
FF + L +PLL L++ QYFVPS FMTP F KEL KMF E + FHHLGRYLFHP
Sbjct: 602 FFTVIIGKLLSLRVPLLILQSDQYFVPSLFMTPFFNKELEKMFVEKDTVFHHLGRYLFHP 781
Query: 247 SNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLGN-----KL-L 300
SNEAW LIT+FYQ +LAKA+E + V N T N KL
Sbjct: 782 SNEAWRLITNFYQAHLAKADEENRSSNKSV-----------QSYFNSTRSNYESSFKLHH 928
Query: 301 PKVLGTKNIYLSSSDKNKK-IMKAVLVSSLYRHYGENLKMTYMNK 344
+ + TK+ + + K+ + VLV+SLY Y +NLK + NK
Sbjct: 929 SQ*VTTKSTWYEKFKRQKQNDCEGVLVASLYPEYXDNLKTXFSNK 1063
>TC84135 similar to SP|Q9M5Q1|FUT1_PEA Galactoside 2-L-fucosyltransferase
(EC 2.4.1.69) (Xyloglucan alpha- (1
2)-fucosyltransferase), partial (41%)
Length = 814
Score = 204 bits (519), Expect = 6e-53
Identities = 106/250 (42%), Positives = 151/250 (60%), Gaps = 6/250 (2%)
Frame = +3
Query: 4 EKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAYN 63
+KLL GLL SGFD+ SC+SR +S K PSPYLIS+LRKYE LH++CGP T +YN
Sbjct: 84 DKLLGGLLASGFDDRSCLSRYRSV---KGLSGNPSPYLISRLRKYEALHKKCGPYTESYN 254
Query: 64 EDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGKDK 123
+ +K + S + CKY++W+ +GLGN+I+++AS+FLYALLT RV+LV G D
Sbjct: 255 KTVKDLNSGH---LSESPDCKYVVWISFSGLGNRILTLASAFLYALLTNRVLLVDPGVDM 425
Query: 124 EGLFCEPFLNSTWLLPEKSPFWNAEKVQTYQSTIKMGGANT---LNEDFPSALHV---NL 177
LFCEP + +W LP P N++ + Q + M N ++ P L + +L
Sbjct: 426 VDLFCEPIPDVSWFLPPDFPL-NSQFLSLNQKSDSMSWKNAKK*ISHGIPKYLLLSTFHL 602
Query: 178 GYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITSTFH 237
+ E++ F CD Q L +P L ++ YF+PS F+ P F +EL+ +FP FH
Sbjct: 603 AHDYDDEDKLFFCDEEQQFLQKVPWLMMKTDNYFIPSLFLMPSFDQELSNLFPNKDKVFH 782
Query: 238 HLGRYLFHPS 247
+GRYLFHP+
Sbjct: 783 FIGRYLFHPT 812
>TC91891 weakly similar to GP|5231145|gb|AAD41092.1| xyloglucan
fucosyltransferase {Arabidopsis thaliana}, partial (20%)
Length = 816
Score = 166 bits (420), Expect(2) = 4e-43
Identities = 78/128 (60%), Positives = 99/128 (76%)
Frame = +3
Query: 4 EKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAYN 63
++ LDGLLVS FDEASC+SR +SH Y K SP+KPS YL+SKLR YE LHR CGPNT+ Y+
Sbjct: 345 DRFLDGLLVSTFDEASCLSRFKSHLYRKASPYKPSAYLLSKLRNYEHLHRSCGPNTKPYS 524
Query: 64 EDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGKDK 123
+ M K K K A T CKY++W +NGLGN++I++A++FLYA+LT RV+LV+FG D
Sbjct: 525 KIMTKGPKFSKG--DADTKCKYLVWTASNGLGNRMITLAAAFLYAILTDRVLLVKFGADM 698
Query: 124 EGLFCEPF 131
GLFCEPF
Sbjct: 699 FGLFCEPF 722
Score = 26.9 bits (58), Expect(2) = 4e-43
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = +1
Query: 127 FCEPFLNSTWLLPEKSPFWNAEKVQTY 153
F F +S+WLLP P+W ++ +TY
Sbjct: 709 FVSLFPDSSWLLPRNFPYW--KRSETY 783
>AJ503783 similar to SP|O22307|C7DB_ Cytochrome P450 71D11 (EC 1.14.-.-)
(Fragment). {Lotus japonicus}, partial (14%)
Length = 320
Score = 30.8 bits (68), Expect = 1.2
Identities = 10/22 (45%), Positives = 15/22 (67%)
Frame = +1
Query: 424 FTLEPCYHYPPLHYCNGKPIED 445
F P +H+PPLH+C+ K E+
Sbjct: 58 FPFHPFFHFPPLHHCDTKTKEE 123
>CA921391 similar to GP|1669591|dbj O-methyltransferase {Glycyrrhiza
echinata}, partial (42%)
Length = 690
Score = 29.3 bits (64), Expect = 3.4
Identities = 14/46 (30%), Positives = 23/46 (49%), Gaps = 6/46 (13%)
Frame = +2
Query: 214 SFFMTPIFKKELNKMFPEITSTFHHLGRYLFHP------SNEAWEL 253
+FF+ PI + +LN + +TF H+ +F P N WE+
Sbjct: 479 AFFIRPIMEHKLNHYSISLWNTFKHISSNMFDPWKGWCMFNHLWEI 616
>TC78840 similar to GP|21689605|gb|AAM67424.1 AT4g27690/T29A15_180
{Arabidopsis thaliana}, partial (64%)
Length = 802
Score = 28.9 bits (63), Expect = 4.4
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = -1
Query: 18 ASCVSRLQSHFYHKPSPH 35
ASCV+ + HF H P PH
Sbjct: 643 ASCVTTINGHFQHIPKPH 590
>BQ144361
Length = 806
Score = 28.5 bits (62), Expect = 5.8
Identities = 28/110 (25%), Positives = 41/110 (36%), Gaps = 4/110 (3%)
Frame = +1
Query: 117 VQFGKDKEGLFCEPFLNSTWLLPEKSPFWNAEKVQTYQSTIKMGGANTLNEDFPSALHVN 176
+ FG+ K G+F P+K FW + G T N P +
Sbjct: 349 ISFGQKKWGVFAHKNFVPHPFFPQKPFFWGGGAPPFLLNRGGCIGGETKNNKTPPPFTNS 528
Query: 177 LGYSPTSE--ERFFHCDHNQFL-LSNIPLLF-LEAGQYFVPSFFMTPIFK 222
LG +P E FF + FL L P F + G+ + FF +F+
Sbjct: 529 LGKNPGKNLGEGFFTPPNGVFLFLKKTPFFFKTKGGEKTLLFFFFIFVFR 678
>CB891069 similar to PIR|E86428|E8 probable ABC transporter [imported] -
Arabidopsis thaliana, partial (17%)
Length = 813
Score = 28.5 bits (62), Expect = 5.8
Identities = 21/87 (24%), Positives = 36/87 (41%), Gaps = 7/87 (8%)
Frame = +1
Query: 309 IYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVYQPSGEEQQKFNDN 368
IY S+ + K M ++ S +Y H+GE++ S + Y + KF DN
Sbjct: 226 IYYQSTSREVKRMDSITRSPVYAHFGESM------NGVSSIRAYKAYDRILHDNGKFMDN 387
Query: 369 -------KHNMKAWVDIYLLSLSDVLV 388
+ W+ I L SL +++
Sbjct: 388 NIRFTLANISTNRWLTIRLESLGGLMI 468
>BG644408 homologue to PIR|T07371|T07 dnaJ protein homolog - potato, partial
(34%)
Length = 582
Score = 28.1 bits (61), Expect = 7.5
Identities = 11/36 (30%), Positives = 17/36 (46%)
Frame = +2
Query: 53 RRCGPNTRAYNEDMKKITKSKKNGTSAATTCKYIIW 88
RRCG ++ M K+ K+ G + C +IW
Sbjct: 377 RRCGGSSNRPKRHMTKMMKTCHGGAQRGSVCTTVIW 484
>TC78573 similar to GP|20197309|gb|AAC63626.2 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 2016
Score = 28.1 bits (61), Expect = 7.5
Identities = 15/39 (38%), Positives = 24/39 (61%)
Frame = +3
Query: 107 YALLTGRVMLVQFGKDKEGLFCEPFLNSTWLLPEKSPFW 145
+ L+ G+ +VQ +D++ L C PFL +T L PE+ W
Sbjct: 903 HPLVKGQSRVVQMYQDQD-LLCIPFLLATVLPPERVAQW 1016
>BG646262 similar to PIR|T06036|T06 hypothetical protein T24A18.10 -
Arabidopsis thaliana, partial (15%)
Length = 677
Score = 27.7 bits (60), Expect = 9.9
Identities = 12/36 (33%), Positives = 20/36 (55%)
Frame = -2
Query: 258 YQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNC 293
YQ +A +E+I L ++ LTP +Q+L+C
Sbjct: 274 YQMTMASNHEKIHLSLQFFSTGLTPLSCYPSQVLSC 167
>TC87728 similar to GP|21554610|gb|AAM63632.1 unknown {Arabidopsis
thaliana}, partial (87%)
Length = 1283
Score = 27.7 bits (60), Expect = 9.9
Identities = 24/112 (21%), Positives = 39/112 (34%), Gaps = 15/112 (13%)
Frame = +3
Query: 1 DEKEKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKP---------------SPYLISKL 45
D K+ + G+DEA S L S HKP P +L+
Sbjct: 180 DFKDAVRQSARSLGYDEAKLASILASFIIHKPPQRSPFTKAAFKTLESIGELDHFLLKHR 359
Query: 46 RKYEELHRRCGPNTRAYNEDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQ 97
+ Y +LHR + +++ K TC+ I + N + N+
Sbjct: 360 KDYTDLHRTTEHERDSIEQEVSAFIK----------TCQEQIDVLKNSINNE 485
>TC85197 GP|1435157|emb|CAA58445.1 histone H3 variant H3.3 {Lycopersicon
esculentum}, complete
Length = 1332
Score = 27.7 bits (60), Expect = 9.9
Identities = 18/58 (31%), Positives = 28/58 (48%)
Frame = -3
Query: 252 ELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLGNKLLPKVLGTKNI 309
EL T NL K+N + + ++P + + L C+LG KLLP+ L T +
Sbjct: 748 ELSTLLILPNLTKSNSSRAVTMWFLNPTSSGCR------LTCSLGCKLLPRCLSTSRL 593
>TC84165 similar to GP|21539497|gb|AAM53301.1 putative protein {Arabidopsis
thaliana}, partial (16%)
Length = 792
Score = 27.7 bits (60), Expect = 9.9
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = +1
Query: 212 VPSFFMTPIFKKELNKMFPEITSTFHHL 239
+P F++ + +NK+FPE T HHL
Sbjct: 508 LPC*FISKHLSQRVNKIFPETIRTHHHL 591
>AJ501178
Length = 630
Score = 27.7 bits (60), Expect = 9.9
Identities = 12/40 (30%), Positives = 24/40 (60%)
Frame = -3
Query: 317 NKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVYQ 356
N I+ + + S+Y +Y +NLK+T ++ T+ ++ YQ
Sbjct: 586 NFLILSKI*ILSIYLYYQQNLKVT*LDVYTIQNKIFLDYQ 467
>TC78781 weakly similar to PIR|T02291|T02291 hypothetical protein T13D8.28 -
Arabidopsis thaliana, partial (12%)
Length = 1300
Score = 27.7 bits (60), Expect = 9.9
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Frame = +3
Query: 380 LLSLSDVLVTT---YQSTFGYVAKALGNSRPWILYNPVYSNEICEREFTLEPCYHYPPLH 436
+LSLS V T + S V++ L R + +P +S IC+RE L PC LH
Sbjct: 561 VLSLSRVQFDTKSVFLSLLSAVSRVLEELR---ISSPTFSAPICDREADLFPCLRGAFLH 731
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,026,012
Number of Sequences: 36976
Number of extensions: 301375
Number of successful extensions: 1714
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 1692
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1707
length of query: 469
length of database: 9,014,727
effective HSP length: 100
effective length of query: 369
effective length of database: 5,317,127
effective search space: 1962019863
effective search space used: 1962019863
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146553.5


