

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.15 - phase: 0
(880 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
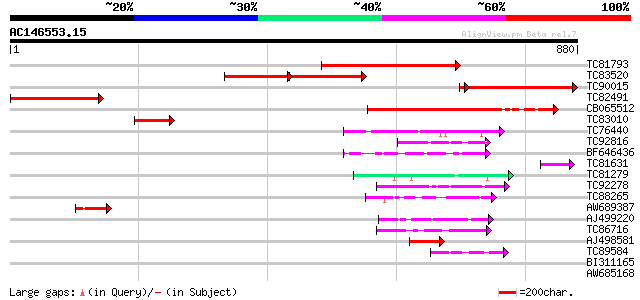
Score E
Sequences producing significant alignments: (bits) Value
TC81793 weakly similar to PIR|D86165|D86165 protein F15K9.3 [imp... 421 e-118
TC83520 weakly similar to PIR|D86165|D86165 protein F15K9.3 [imp... 233 e-114
TC90015 similar to PIR|T01393|T01393 apoptosis inhibitor homolog... 335 1e-94
TC82491 283 2e-76
CB065512 similar to GP|3608154|gb| unknown protein {Arabidopsis ... 225 7e-59
TC83010 similar to GP|12322447|gb|AAG51242.1 unknown protein 3'... 62 7e-10
TC76440 weakly similar to SP|Q9LD55|IF3A_ARATH Eukaryotic transl... 55 9e-08
TC92816 weakly similar to GP|18076679|emb|CAC84774. P70 protein ... 49 1e-05
BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis th... 48 1e-05
TC81631 similar to GP|20279471|gb|AAM18751.1 unknown protein {Or... 47 4e-05
TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.... 47 4e-05
TC92278 similar to SP|O43093|KINH_SYNRA Kinesin heavy chain (Syn... 47 4e-05
TC88265 weakly similar to GP|18176104|gb|AAL59984.1 unknown prot... 45 9e-05
AW689387 similar to GP|10727986|gb CG18331 gene product {Drosoph... 45 1e-04
AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c... 45 1e-04
TC86716 similar to PIR|T14279|T14279 myosin-like protein my5 - c... 44 4e-04
AJ498581 44 4e-04
TC89584 weakly similar to GP|18076679|emb|CAC84774. P70 protein ... 42 8e-04
BI311165 similar to GP|16902294|d kinesin-related protein {Arabi... 42 0.001
AW685168 weakly similar to PIR|T01757|T0 hypothetical protein A_... 41 0.002
>TC81793 weakly similar to PIR|D86165|D86165 protein F15K9.3 [imported] -
Arabidopsis thaliana, partial (16%)
Length = 752
Score = 421 bits (1082), Expect = e-118
Identities = 215/215 (100%), Positives = 215/215 (100%)
Frame = +3
Query: 485 SLSSKGNSSIAPICCSNKSHSSSCVGIPYDKSMRQWLPQDRKDELILKMVPRVRELQNEL 544
SLSSKGNSSIAPICCSNKSHSSSCVGIPYDKSMRQWLPQDRKDELILKMVPRVRELQNEL
Sbjct: 6 SLSSKGNSSIAPICCSNKSHSSSCVGIPYDKSMRQWLPQDRKDELILKMVPRVRELQNEL 185
Query: 545 QEWTEWANQKVMQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENAL 604
QEWTEWANQKVMQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENAL
Sbjct: 186 QEWTEWANQKVMQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENAL 365
Query: 605 GKAGGQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWE 664
GKAGGQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWE
Sbjct: 366 GKAGGQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWE 545
Query: 665 NQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFE 699
NQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFE
Sbjct: 546 NQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFE 650
>TC83520 weakly similar to PIR|D86165|D86165 protein F15K9.3 [imported] -
Arabidopsis thaliana, partial (5%)
Length = 773
Score = 233 bits (594), Expect(2) = e-114
Identities = 115/120 (95%), Positives = 116/120 (95%)
Frame = +1
Query: 435 GPSTPTFSLDSSDTISRAADSSSSEHEANLIPAVSSPPDALSATDTDLSLSLSSKGNSSI 494
GPSTPTFSLDSSDTISRAADSSSSEHEANLIPAVSSPPDALSATDTDLSLSLSSKGNSSI
Sbjct: 307 GPSTPTFSLDSSDTISRAADSSSSEHEANLIPAVSSPPDALSATDTDLSLSLSSKGNSSI 486
Query: 495 APICCSNKSHSSSCVGIPYDKSMRQWLPQDRKDELILKMVPRVRELQNELQEWTEWANQK 554
APICCSNKSHSSSCVGIPYDKSMRQWLPQDRKDELILKM PRVRELQNELQEW WAN +
Sbjct: 487 APICCSNKSHSSSCVGIPYDKSMRQWLPQDRKDELILKMGPRVRELQNELQEWXXWANXR 666
Score = 199 bits (507), Expect(2) = e-114
Identities = 102/106 (96%), Positives = 103/106 (96%)
Frame = +3
Query: 334 SPLLQEKCGIVRKVHSSSTKREYIFRQKSIHVEKSYRTYGSKGSSRGGKLSGLSGLILDK 393
SPLLQEKCGIVRKVHSSSTKREYIFRQKSIHVEKSYRTYGSKGSSRGGKLSGLSGLIL+K
Sbjct: 3 SPLLQEKCGIVRKVHSSSTKREYIFRQKSIHVEKSYRTYGSKGSSRGGKLSGLSGLILNK 182
Query: 394 KLKSVSESTAINLKSASINISKAVGIDVTQNNHNTHFSSNNGPSTP 439
KLKSVSESTAINLKSASINISKAVGIDVTQNNHNTHFSSNNG P
Sbjct: 183 KLKSVSESTAINLKSASINISKAVGIDVTQNNHNTHFSSNNGTIYP 320
>TC90015 similar to PIR|T01393|T01393 apoptosis inhibitor homolog T4I9.12 -
Arabidopsis thaliana, partial (8%)
Length = 757
Score = 335 bits (860), Expect(2) = 1e-94
Identities = 168/174 (96%), Positives = 170/174 (97%)
Frame = +1
Query: 707 KKTEELLSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKS 766
+K + SMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKS
Sbjct: 94 RKQKNFFSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKS 273
Query: 767 DSSKIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISELVQKLNNFSMNGGGVKREREC 826
DSSKIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISELVQKLNNFSMNGGGVKREREC
Sbjct: 274 DSSKIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISELVQKLNNFSMNGGGVKREREC 453
Query: 827 VMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVRYART 880
VMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVRYART
Sbjct: 454 VMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVRYART 615
Score = 30.4 bits (67), Expect(2) = 1e-94
Identities = 14/15 (93%), Positives = 15/15 (99%)
Frame = +3
Query: 699 EAKRRQAAKKTEELL 713
+AKRRQAAKKTEELL
Sbjct: 69 QAKRRQAAKKTEELL 113
>TC82491
Length = 678
Score = 283 bits (725), Expect = 2e-76
Identities = 144/145 (99%), Positives = 145/145 (99%)
Frame = +2
Query: 1 MASLVVSGNSQMSSSVSVQEKGSRNKRKFRADPPLGESSKSISSLQHESLSYEFSAEKVE 60
MASLVVSGNSQMSSSVSVQEKGSRNKRKFRADPPLGESSKSISSLQHESLSYEFSAEKVE
Sbjct: 242 MASLVVSGNSQMSSSVSVQEKGSRNKRKFRADPPLGESSKSISSLQHESLSYEFSAEKVE 421
Query: 61 ITPCFGPVTASDLCSVSHGCSDGLKLDLGLSSPAVSSEVRLCQPKEELEVVESHGADWSD 120
ITPCFGPVTASDLCSVSHGCSDGLKLDLGLSSPAVSSEVRLCQPKEELEVVESHGADWSD
Sbjct: 422 ITPCFGPVTASDLCSVSHGCSDGLKLDLGLSSPAVSSEVRLCQPKEELEVVESHGADWSD 601
Query: 121 HTETQLQELVLSNLQTIFKSAIKKI 145
HTETQLQELVLSNLQTIFKSAIKK+
Sbjct: 602 HTETQLQELVLSNLQTIFKSAIKKL 676
>CB065512 similar to GP|3608154|gb| unknown protein {Arabidopsis thaliana},
partial (34%)
Length = 852
Score = 225 bits (573), Expect = 7e-59
Identities = 127/297 (42%), Positives = 192/297 (63%)
Frame = +1
Query: 556 MQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERAN 615
+QAA++LS D ELKTLR ++EE ++LKK KQ LE+ TMK+LSEMENAL KA GQV+RAN
Sbjct: 1 LQAAKKLSSDLTELKTLRMDREETQKLKKGKQALEDTTMKRLSEMENALRKASGQVDRAN 180
Query: 616 TAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEELM 675
AVR+LE ENA +R EMEA+KL A ES T EV+K+EKK K +WE QK+ LQ+E+
Sbjct: 181 GAVRRLETENAEIRAEMEASKLSASESVTACLEVAKKEKKYLKKLLAWEKQKAKLQKEIS 360
Query: 676 TEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRKEREQIEELARTKEE 735
K K+ + S Q + + ++ EAK ++ K E+ L++V R+ +E E + E
Sbjct: 361 DLKEKILEDREVSAQNKQRQKEAEAKWKEELKAQEDALALVDEERRSKEAAESDNKRGFE 540
Query: 736 RIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGIDGSYAGSFKDTKKGSGF 795
++L+ E + +R+KDD+ +LE ++++++ S + AAL + S F+ TK
Sbjct: 541 ALRLKIELDFQRHKDDLSRLENDLSRLKA---SVRSAALHHQ-NTSPIKDFEGTK----- 693
Query: 796 EEPHTASISELVQKLNNFSMNGGGVKRERECVMCLSEEMSVVFLPCAHQVVCTKCNE 852
P +I++L+ L++ S + REC++C+ +E+SVVFLPCAH V+C KC++
Sbjct: 694 --PQRETIAKLLLDLDDLSES--EANNNRECIICMKDEVSVVFLPCAHPVMCAKCSD 852
>TC83010 similar to GP|12322447|gb|AAG51242.1 unknown protein 3' partial;
1386-1 {Arabidopsis thaliana}, partial (18%)
Length = 659
Score = 62.4 bits (150), Expect = 7e-10
Identities = 31/63 (49%), Positives = 41/63 (64%)
Frame = +3
Query: 194 EHYFKDLAELQNYILAELVCVLQEVRPFFSFGDAMWCLLISDMNVSHACAMDGDPLSSLG 253
E F DL L+ Y LA +VC+LQ+VRP S GDAMWCLL+SD++V A ++ S G
Sbjct: 24 EPVFADLKHLEEYSLAGMVCLLQQVRPNLSKGDAMWCLLMSDLHVGKASTIEIPVPGSAG 203
Query: 254 SDG 256
+ G
Sbjct: 204 ARG 212
>TC76440 weakly similar to SP|Q9LD55|IF3A_ARATH Eukaryotic translation
initiation factor 3 subunit 10 (eIF-3 theta), partial
(41%)
Length = 1953
Score = 55.5 bits (132), Expect = 9e-08
Identities = 67/277 (24%), Positives = 132/277 (47%), Gaps = 27/277 (9%)
Frame = +1
Query: 518 RQWLPQDRKDELILKMVPRVRELQNE---LQEWTEWANQKVMQAARRLSKDKAELKTLRQ 574
R+ + + RK+E +++ + RE +++ L + E A Q RRL+ + + K R
Sbjct: 418 RKSIIEKRKEEQERQLLEKEREEESKRLRLLKIDEEAEQ------RRLATEIEQRKIQRL 579
Query: 575 EKEEVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERANTAVRKLEMENAALRKEMEA 634
++E+ ER ++E + L K+L GGQ+ R + L +E R+EME
Sbjct: 580 QREKEERDREEAEALRLEAEKRLKRKGKKPVIEGGQISRESLMQLTL-VEQVRERQEMEK 756
Query: 635 AKLRAVESATNFQEVSKREKKTQMKFQSWENQ----------KSLLQEEL--------MT 676
KL+ + + E +KRE+ + +++ + + LQ EL +
Sbjct: 757 -KLQKLAKTMDHLERAKREEAAPLIDAAYQQRLVEERVLHEREQQLQVELSRQRHAGDLN 933
Query: 677 EKNKLAHI--SKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRKEREQIEELAR--- 731
EK +L+ + +KE Q V + + R + + + ++ S ++ERE++ +L
Sbjct: 934 EKERLSRMMGNKEIYQERVVSHRQAEFNRLQRDRLDRISKILLSRKQEREKMRKLKYYLK 1113
Query: 732 -TKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSD 767
+E+R KL E+E R +++ ++ +KE A+ + K D
Sbjct: 1114VEEEKRQKLREEEEARE-REEAERKKKEQAERQAKLD 1221
>TC92816 weakly similar to GP|18076679|emb|CAC84774. P70 protein {Nicotiana
tabacum}, partial (21%)
Length = 628
Score = 48.5 bits (114), Expect = 1e-05
Identities = 41/144 (28%), Positives = 70/144 (48%)
Frame = +3
Query: 603 ALGKAGGQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQS 662
A + Q++ N+ +RK + R E + +S EKK K Q
Sbjct: 222 ATPRTARQLKTPNSGSNSASSSPNPIRKTPKDMSPRVNERRLSHSPIS--EKKRPSKVQE 395
Query: 663 WENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRKE 722
E+Q + LQE+L + K++L S ES + +VQ E EAK+ ++LS+ + +
Sbjct: 396 LESQIAKLQEDLKSAKDQLN--SSESWKRKVQEEAEEAKK--------QILSLSKELEES 545
Query: 723 REQIEELARTKEERIKLEAEKELR 746
R+Q +L+ ++E R++ E K LR
Sbjct: 546 RQQFSDLSASEETRLQ-ELSKYLR 614
>BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis thaliana},
partial (11%)
Length = 659
Score = 48.1 bits (113), Expect = 1e-05
Identities = 53/228 (23%), Positives = 102/228 (44%)
Frame = +1
Query: 518 RQWLPQDRKDELILKMVPRVRELQNELQEWTEWANQKVMQAARRLSKDKAELKTLRQEKE 577
R+W DEL LK +EL+N LQ+ K E+ L+Q +E
Sbjct: 133 REW------DELDLKRADVEKELKNVLQQ-------------------KEEILKLQQNEE 237
Query: 578 EVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERANTAVRKLEMENAALRKEMEAAKL 637
E RLKKEKQ E+ ++L ++ A + ++E+E ++L ++ + K
Sbjct: 238 E--RLKKEKQATEDYLQRELETLQLA----------KESFAAEMELEKSSLAEKAQNEKN 381
Query: 638 RAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQ 697
+ + E+ ++E + M+ Q + +K L + + E E +++E+
Sbjct: 382 QLLLDF----EMRRKELEADMQNQLEQKEKDLFESRRLFE---------EKRESELNNIN 522
Query: 698 FEAKRRQAAKKTEELLSMVSSIRKEREQIEELARTKEERIKLEAEKEL 745
F R A + EE+ S + +E++ +E R ER ++E ++++
Sbjct: 523 F--LREVANRGMEEMKHQRSKLEREKQDADE-NRKHVERQRIEMQEDI 657
>TC81631 similar to GP|20279471|gb|AAM18751.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (40%)
Length = 787
Score = 46.6 bits (109), Expect = 4e-05
Identities = 18/53 (33%), Positives = 30/53 (55%)
Frame = +1
Query: 824 RECVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVR 876
+ECV+CLSE + PC H +C+ C ++ Q CP CR P++ + ++
Sbjct: 415 KECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQ-TNRCPICRQPVERLLEIK 570
>TC81279 similar to PIR|T47949|T47949 hypothetical protein F2A19.170 -
Arabidopsis thaliana, partial (31%)
Length = 1205
Score = 46.6 bits (109), Expect = 4e-05
Identities = 60/295 (20%), Positives = 110/295 (36%), Gaps = 46/295 (15%)
Frame = +2
Query: 534 VPRVRELQNELQEWTEWANQKVMQAARRLSKD-KAELKTLRQEKEEVERLKKEKQCLEEN 592
V ++ LQ EL M +++RL + + E TL+ EK E+E K + +
Sbjct: 179 VSEIKRLQLELTRQRSKEASNAMDSSKRLIETLEKENTTLKMEKSELEAAVKASSASDLS 358
Query: 593 TMKK-----------LSEMENALGKAGGQVERANTAVRKL-----EMENAALRKEMEAAK 636
K L +M N L K + ++A + +L E EN K E K
Sbjct: 359 DPSKSFPGKEDMEISLQKMSNDLKKTQQERDKAVQELTRLKQHLLEKENEESEKMDEDTK 538
Query: 637 -LRAVESATNF--QEVSKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAEV 693
+ + + N+ ++S E+ + E KS E++T + + ++K+
Sbjct: 539 VIEELRDSNNYLRAQISHLERALEQATSDQEKLKSANNSEILTSREVIDDLNKKLTNC-- 712
Query: 694 QAEQFEAKRRQAAKKTEELLSMVSSIRKEREQIEELARTKEERIKL-------------- 739
+AK + L + I + EELAR +EE L
Sbjct: 713 -ISTIDAKNIELINLQTALGQYYAEIEAKEHLEEELARAREETANLSQLLKDADSRVDIL 889
Query: 740 ------------EAEKELRRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGIDGSY 782
++EK ++ + KLE+ A++R+ + S + +D +
Sbjct: 890 SGEKEEILAKLSQSEKVQSEWRSRVSKLEERNAKLRRALEQSMTRLNRMSVDSDF 1054
Score = 29.6 bits (65), Expect = 5.3
Identities = 31/149 (20%), Positives = 62/149 (40%)
Frame = +2
Query: 662 SWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRK 721
+W+ QEE K+ +E K +++ ++ + RQ K+ E +S + ++
Sbjct: 38 NWKTFSLQFQEEQKLNKS----FQEELKLLKLERDKTTTEVRQLHKELNEKVSEIKRLQL 205
Query: 722 EREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGIDGS 781
E + R+KE +++ K L I+ LEKE ++ + + L+ + S
Sbjct: 206 ELTR----QRSKEASNAMDSSKRL------IETLEKENTTLKMEK-----SELEAAVKAS 340
Query: 782 YAGSFKDTKKGSGFEEPHTASISELVQKL 810
A D K +E S+ ++ L
Sbjct: 341 SASDLSDPSKSFPGKEDMEISLQKMSNDL 427
>TC92278 similar to SP|O43093|KINH_SYNRA Kinesin heavy chain (Synkin).
{Syncephalastrum racemosum}, partial (10%)
Length = 709
Score = 46.6 bits (109), Expect = 4e-05
Identities = 44/209 (21%), Positives = 98/209 (46%), Gaps = 3/209 (1%)
Frame = +3
Query: 570 KTLRQEKEEVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERANTAVRKLEMENAALR 629
K +Q ++ + +L+ TM++LS + L ++ V + + +L EN L
Sbjct: 60 KKEQQIRDTLLKLETISDTSSPLTMEELSTLRRELSESKSLVTQHEQTINELHYENEHLT 239
Query: 630 KEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESK 689
++ + ++R ++E+ KT + ++ N + E + + KL ++
Sbjct: 240 RKRDELEIRLTTLELEYEELLD---KTIAEEEA--NNNIDITETIAELRGKLE--AQFVA 398
Query: 690 QAEVQAEQFEAKRRQAAKKTEE---LLSMVSSIRKEREQIEELARTKEERIKLEAEKELR 746
+ EVQ ++ + +++ KK EE L S ++ ++K E ++ KE IK + +K +
Sbjct: 399 KKEVQQKEIDELKQELEKKNEELHKLNSALADLKKVNEDLQTAINDKE--IKSDGQKNVA 572
Query: 747 RYKDDIQKLEKEIAQIRQKSDSSKIAALK 775
+ D++++ K +AQ D K A ++
Sbjct: 573 DKEKDMERIXKTMAQQLADFDVMKKALMR 659
>TC88265 weakly similar to GP|18176104|gb|AAL59984.1 unknown protein
{Arabidopsis thaliana}, partial (65%)
Length = 1632
Score = 45.4 bits (106), Expect = 9e-05
Identities = 57/216 (26%), Positives = 102/216 (46%), Gaps = 13/216 (6%)
Frame = +3
Query: 553 QKVMQAARRLSKDKAELKTLRQEK---EEV-----ERLKKEKQCLEENTMKKLSEMENAL 604
Q + A+ + K+ LKT+++ K E+V E L +CL++ + E + A+
Sbjct: 246 QDTLNLAKGMQKELEMLKTIKESKGPLEDVTHGSNENLLSFMKCLKDRGVS--IETQEAM 419
Query: 605 GKAGGQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWE 664
VE AN+ + KL+ + R + A+ ++E S K T +S
Sbjct: 420 A-----VEEANSLMLKLKAQLEPFRY--------VADEASPWEEKSAVAKFTNKVHKS-- 554
Query: 665 NQKSLLQEELMTEKNKLAHISKESKQAEVQA---EQFEAKRRQAAK-KTEELLSMVSSIR 720
++NKL K + AE+ A EQF+ R+A + + E+ ++SI+
Sbjct: 555 ------------KRNKLWRKKKRKRIAEMLAKEHEQFDQIDREADEWRAREIAKEIASIK 698
Query: 721 -KEREQIEELARTKEERIKLEAEKELRRYKDDIQKL 755
K+ +I EL + KEE+ KLE+E EL ++ +++L
Sbjct: 699 VKKMNEIGEL-KVKEEKKKLESELELLLMEEKLKEL 803
>AW689387 similar to GP|10727986|gb CG18331 gene product {Drosophila
melanogaster}, partial (1%)
Length = 613
Score = 45.1 bits (105), Expect = 1e-04
Identities = 24/58 (41%), Positives = 38/58 (65%), Gaps = 1/58 (1%)
Frame = +1
Query: 102 CQPKEELEVVESHGADWSDHTETQLQELVLSNLQTIFKSAIKKIVAC-GYTEDVATKA 158
C +++L V+E DW D TQL++L++SNL+ IF +AIK V GY++++A A
Sbjct: 445 CTEEQQLGVLEVE--DWKDPMATQLEDLLMSNLEAIFSNAIKXXVDLGGYSQEMAEMA 612
>AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c in
chromosome I. [Fission yeast] {Schizosaccharomyces
pombe}, partial (12%)
Length = 518
Score = 45.1 bits (105), Expect = 1e-04
Identities = 45/178 (25%), Positives = 77/178 (42%)
Frame = -3
Query: 573 RQEKEEVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERANTAVRKLEMENAALRKEM 632
R ++EE E + +E+ + E E G+ Q E ME ++
Sbjct: 495 RMQREEDESAMDRGEYYDEDEDEDEDEYEED-GEEDNQTEEQR-------MEEGRRMFQI 340
Query: 633 EAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAE 692
AA++ ++E +E+ Q K +++ L+EE +K K K +A
Sbjct: 339 FAARMFEQRVLNAYREKVAQER--QQKLLEELEEENRLKEERELKKLKEKERKKAKNRAL 166
Query: 693 VQAEQFEAKRRQAAKKTEELLSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKD 750
Q ++ E RR+A + EE +E++ EE R +EER+K EAE+ R+ D
Sbjct: 165 KQQKEEERARREAERLAEE---KAQRAEREKKLEEERKRREEERLKKEAERRQRKKND 1
>TC86716 similar to PIR|T14279|T14279 myosin-like protein my5 - common
sunflower, partial (38%)
Length = 2675
Score = 43.5 bits (101), Expect = 4e-04
Identities = 53/184 (28%), Positives = 87/184 (46%), Gaps = 6/184 (3%)
Frame = +2
Query: 570 KTLRQEKEEVERLKKEKQC---LEENTMKKLSEMENALGKAGGQVERANTAVRKLEMENA 626
K ++ +E RL+ EK+ LEE ++++++ +AL QVE AN A
Sbjct: 2 KLEKRVEELTWRLQIEKRLRTDLEEEKAQEVAKLRDALHAMQIQVEEAN----------A 151
Query: 627 ALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISK 686
+ KE EA S Q+ K+T + + E SLL E + K
Sbjct: 152 KVIKEREA-------SQKAIQDAPPVIKETPVIIEDTEKINSLLAE---------VNCLK 283
Query: 687 ESKQAEVQAEQFEAKRRQA--AKKTEELLSMVSSIRKEREQIEELARTKEERI-KLEAEK 743
ES E +A++ EAKR QA +++EL V ++ +Q++EL + EE+I E+E
Sbjct: 284 ESLLLEREAKE-EAKRAQAETEARSKELFKKVEDSDRKADQLQELVQRLEEKISNSESEN 460
Query: 744 ELRR 747
++ R
Sbjct: 461 QVLR 472
>AJ498581
Length = 613
Score = 43.5 bits (101), Expect = 4e-04
Identities = 21/54 (38%), Positives = 35/54 (63%)
Frame = -3
Query: 621 LEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEEL 674
LE + + L KE+ +AK A +S ++Q+ +RE+ T + QSW+ + LLQ+EL
Sbjct: 605 LEAKKSLLEKELVSAKSLAEKSMASYQQALEREQTTIQQAQSWKTELDLLQDEL 444
>TC89584 weakly similar to GP|18076679|emb|CAC84774. P70 protein {Nicotiana
tabacum}, partial (26%)
Length = 872
Score = 42.4 bits (98), Expect = 8e-04
Identities = 37/124 (29%), Positives = 61/124 (48%), Gaps = 3/124 (2%)
Frame = +3
Query: 653 EKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEEL 712
EK + E++ S L+E+L K +L S ES + + Q E +AK R L
Sbjct: 441 EKNGTNRVHELESRLSQLEEDLKKAKEQLN--SSESLRIKAQHEAEDAKNR--------L 590
Query: 713 LSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQK---LEKEIAQIRQKSDSS 769
LSM + + + Q+ EL+ +++ERI +ELR+ D + E E Q + DS+
Sbjct: 591 LSMSRELEESQRQLLELSNSEDERI-----QELRKISQDRDREWQSELEAVQKQHSMDSA 755
Query: 770 KIAA 773
+A+
Sbjct: 756 ALAS 767
>BI311165 similar to GP|16902294|d kinesin-related protein {Arabidopsis
thaliana}, partial (16%)
Length = 657
Score = 42.0 bits (97), Expect = 0.001
Identities = 44/188 (23%), Positives = 73/188 (38%), Gaps = 14/188 (7%)
Frame = +2
Query: 549 EWANQKVMQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENALGKAG 608
E N+K+ LS+D + L+ Q+ E KE +K L+ L
Sbjct: 17 EIENEKLKLEQVHLSEDNSGLRVQNQKLSEEASYAKELASAAAVELKNLAGEVTKLSLQN 196
Query: 609 GQVERANTAVRKL-------EMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQ 661
++E+ VR L +M N RK +A R ++ ++S F
Sbjct: 197 AKLEKELMTVRDLANSRVAVQMVNGVNRKYSDARSGRKGRISSRANDLSGAGLDD---FD 367
Query: 662 SWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLS------- 714
SW L+ EL K + A + + E E+F K +A K+ E L +
Sbjct: 368 SWSLDADDLRLELQARKQREAALESALSEKEFVEEEFRKKAEEAKKREEALENDLANMWV 547
Query: 715 MVSSIRKE 722
+V+ ++KE
Sbjct: 548 LVAKLKKE 571
>AW685168 weakly similar to PIR|T01757|T0 hypothetical protein A_IG002P16.20
- Arabidopsis thaliana, partial (17%)
Length = 607
Score = 40.8 bits (94), Expect = 0.002
Identities = 47/242 (19%), Positives = 101/242 (41%), Gaps = 5/242 (2%)
Frame = +2
Query: 552 NQKVMQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENALGKAGGQV 611
N K R + +ELK LR RL ++Q + T+ +
Sbjct: 26 NDKKQSFRRNVVSLASELKELR------SRLASQEQSYVKETLTR--------------- 142
Query: 612 ERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQ 671
+ A T + +E+E L+K++E EK Q++ + +K + +
Sbjct: 143 QEAETNAKVMELEIGKLQKKLE-------------------EKNEQLQVSTSSAEKHIKE 265
Query: 672 EELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRKEREQIEELAR 731
+ + + + + +S A Q+ Q + Q +EL SS+R+ E++ L
Sbjct: 266 LDDLRTQLVITRATADSSAASTQSSQLQCFELQ-----KELDEKNSSLREHEERVIRLGE 430
Query: 732 -----TKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGIDGSYAGSF 786
K+ + + ++K+LR D++ ++E++I + KS +K L + +D ++ +
Sbjct: 431 QLDNLQKDLKSRESSQKQLR---DEVFRIERDIMEALAKSGENKDCELXKILDEVFS*EY 601
Query: 787 KD 788
++
Sbjct: 602 RE 607
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.125 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,270,064
Number of Sequences: 36976
Number of extensions: 340843
Number of successful extensions: 2092
Number of sequences better than 10.0: 152
Number of HSP's better than 10.0 without gapping: 1983
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2055
length of query: 880
length of database: 9,014,727
effective HSP length: 105
effective length of query: 775
effective length of database: 5,132,247
effective search space: 3977491425
effective search space used: 3977491425
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146553.15


