

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.13 + phase: 0
(198 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
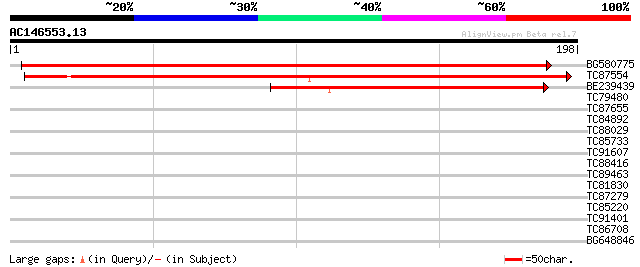
Score E
Sequences producing significant alignments: (bits) Value
BG580775 weakly similar to GP|15529149|gb AT3g21550/MIL23_11 {Ar... 377 e-105
TC87554 weakly similar to GP|15529149|gb|AAK97669.1 AT3g21550/MI... 258 1e-69
BE239439 similar to GP|9757729|dbj| gene_id:MCL19.14~pir||T09894... 88 2e-18
TC79480 30 0.80
TC87655 similar to GP|4587225|dbj|BAA36171.1 FEN-1 {Oryza sativa... 30 0.80
TC84892 homologue to GP|4836403|gb|AAD30426.1| seed maturation p... 29 1.4
TC88029 similar to PIR|T52119|T52119 signal peptidase (EC 3.4.99... 27 5.2
TC85733 similar to GP|3832512|gb|AAC70779.1| granule-bound glyco... 27 5.2
TC91607 similar to GP|13548330|emb|CAC35877. putative protein {A... 27 6.8
TC88416 similar to GP|21703136|gb|AAM74508.1 AT4g32250/F10M6_110... 27 6.8
TC89463 weakly similar to GP|13447813|gb|AAK26764.1 small basic ... 27 6.8
TC81830 similar to GP|15982206|emb|CAC83608. Na+/H+ antiporter ... 27 6.8
TC87279 weakly similar to GP|8843755|dbj|BAA97303.1 gene_id:MXK3... 26 8.8
TC85220 weakly similar to PIR|S33520|S33520 Lea protein - soybea... 26 8.8
TC91401 similar to PIR|H96582|H96582 F15I1.27 [imported] - Arabi... 26 8.8
TC86708 similar to GP|21553407|gb|AAM62500.1 unknown {Arabidopsi... 26 8.8
BG648846 weakly similar to GP|8843755|dbj| gene_id:MXK3.14~unkno... 26 8.8
>BG580775 weakly similar to GP|15529149|gb AT3g21550/MIL23_11 {Arabidopsis
thaliana}, partial (87%)
Length = 557
Score = 377 bits (969), Expect = e-105
Identities = 185/185 (100%), Positives = 185/185 (100%)
Frame = +1
Query: 5 ASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSG 64
ASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSG
Sbjct: 1 ASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSG 180
Query: 65 ILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHA 124
ILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHA
Sbjct: 181 ILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHA 360
Query: 125 FLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
FLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY
Sbjct: 361 FLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 540
Query: 185 PTSSD 189
PTSSD
Sbjct: 541 PTSSD 555
>TC87554 weakly similar to GP|15529149|gb|AAK97669.1 AT3g21550/MIL23_11
{Arabidopsis thaliana}, partial (85%)
Length = 1007
Score = 258 bits (658), Expect = 1e-69
Identities = 122/192 (63%), Positives = 153/192 (79%), Gaps = 1/192 (0%)
Frame = +2
Query: 6 SSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGI 65
S+ T T ST+ K+ G+G+L+KLLPTGTVFLFQ+L+PVVTN+G C T NKYLS I
Sbjct: 95 STFNTETTTATSTIK-KSIGGIGSLIKLLPTGTVFLFQFLNPVVTNSGRCKTSNKYLSSI 271
Query: 66 LLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPS-PGSDSVDLSAYKLRFGDFVHA 124
LLVICGFNC F++FTDSYTG+D +RHYGIVT GLWPS P S S+DL+ Y+L+ DFVHA
Sbjct: 272 LLVICGFNCFFSTFTDSYTGTDKKRHYGIVTTKGLWPSPPASTSIDLTKYRLKGSDFVHA 451
Query: 125 FLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
LS+++FA+LGLLDTN VHCFYP FES++K L+QVLPP IGV G +F++FP +RHGIGY
Sbjct: 452 ALSLLIFALLGLLDTNTVHCFYPSFESTQKQLLQVLPPTIGVFVGWMFVMFPQHRHGIGY 631
Query: 185 PTSSDTNDTSEK 196
P S+D + + K
Sbjct: 632 PVSTDDSHDASK 667
>BE239439 similar to GP|9757729|dbj| gene_id:MCL19.14~pir||T09894~similar to
unknown protein {Arabidopsis thaliana}, partial (32%)
Length = 393
Score = 87.8 bits (216), Expect = 2e-18
Identities = 40/100 (40%), Positives = 62/100 (62%), Gaps = 3/100 (3%)
Frame = +2
Query: 92 YGIVTMNGLWPSPGSDSVD---LSAYKLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPK 148
+G T GLW GS + + Y+++F DF+HA +S++VFA + L D NVV+CF+P+
Sbjct: 5 HGFATFKGLWVIDGSTKLPPQVAAKYRIKFIDFMHAMMSILVFAAIALFDQNVVNCFFPE 184
Query: 149 FESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSS 188
+ ++ LP IGV +F+ FP+ RHGIG+P S+
Sbjct: 185 PSKEIQEILTALPVAIGVFCSMLFVAFPTERHGIGFPLST 304
>TC79480
Length = 644
Score = 29.6 bits (65), Expect = 0.80
Identities = 17/50 (34%), Positives = 29/50 (58%)
Frame = -3
Query: 29 NLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTS 78
+L+ +LP G V +F ++ P++TN+ +N + I L+ FNC TS
Sbjct: 507 HLIAILPKGIVSMFFHIKPIITNH---*MVNHH---IFLMSTRFNCFPTS 376
>TC87655 similar to GP|4587225|dbj|BAA36171.1 FEN-1 {Oryza sativa (japonica
cultivar-group)}, partial (92%)
Length = 1120
Score = 29.6 bits (65), Expect = 0.80
Identities = 10/29 (34%), Positives = 17/29 (58%)
Frame = -1
Query: 51 NNGHCSTINKYLSGILLVICGFNCAFTSF 79
NNGH K + +++++C +C FT F
Sbjct: 502 NNGHSHESQKSFAVVIMLLCHLHCPFTEF 416
>TC84892 homologue to GP|4836403|gb|AAD30426.1| seed maturation protein PM27
{Glycine max}, partial (20%)
Length = 481
Score = 28.9 bits (63), Expect = 1.4
Identities = 11/21 (52%), Positives = 16/21 (75%)
Frame = +2
Query: 55 CSTINKYLSGILLVICGFNCA 75
CS I +LSG+L++ICG C+
Sbjct: 395 CSLICTWLSGVLVMICGDTCS 457
>TC88029 similar to PIR|T52119|T52119 signal peptidase (EC 3.4.99.-)
[imported] - Arabidopsis thaliana, partial (98%)
Length = 764
Score = 26.9 bits (58), Expect = 5.2
Identities = 15/47 (31%), Positives = 22/47 (45%), Gaps = 1/47 (2%)
Frame = +3
Query: 67 LVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSP-GSDSVDLS 112
+ I GF CA SFTD+ ++ +N P G+D V L+
Sbjct: 45 VTILGFICAIASFTDTLNSPSPSVQVQVLNVNWFQKQPNGNDEVYLT 185
>TC85733 similar to GP|3832512|gb|AAC70779.1| granule-bound glycogen
(starch) synthase {Astragalus membranaceus}, partial
(94%)
Length = 2251
Score = 26.9 bits (58), Expect = 5.2
Identities = 20/58 (34%), Positives = 25/58 (42%), Gaps = 8/58 (13%)
Frame = +1
Query: 5 ASSSKTSTNKGASTMTGKTFSGVGNLVKL--------LPTGTVFLFQYLSPVVTNNGH 54
+SSSKTS G+ GK G NLV + G + L PV+ NGH
Sbjct: 211 SSSSKTSEKSGSGKFNGKIVCGGMNLVFVGAEVGPWSKTGGLGDVLGGLPPVLAGNGH 384
>TC91607 similar to GP|13548330|emb|CAC35877. putative protein {Arabidopsis
thaliana}, partial (9%)
Length = 636
Score = 26.6 bits (57), Expect = 6.8
Identities = 18/60 (30%), Positives = 25/60 (41%), Gaps = 2/60 (3%)
Frame = +1
Query: 2 SEKASSSKTSTNKGASTMTGKTFSGVGNLVKLLPT--GTVFLFQYLSPVVTNNGHCSTIN 59
+ ASSS T T + F L PT +F+F+ L P + HCS +N
Sbjct: 97 NNSASSSHTHTYFNDHCLQSFFFKS*PTFSTLFPTFQTIIFIFESLFPSPLSQEHCSLLN 276
>TC88416 similar to GP|21703136|gb|AAM74508.1 AT4g32250/F10M6_110
{Arabidopsis thaliana}, partial (52%)
Length = 2146
Score = 26.6 bits (57), Expect = 6.8
Identities = 24/77 (31%), Positives = 32/77 (41%), Gaps = 3/77 (3%)
Frame = -1
Query: 25 SGVGNLVKLLPTGTV---FLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFTD 81
S +L+K T T+ FL Y + H I G L ICGF+C T TD
Sbjct: 955 SSFSDLLKARRTSTIKGRFLISY-------SKHALRIFSMGEGRPLGICGFSCFST--TD 803
Query: 82 SYTGSDGQRHYGIVTMN 98
Y G + G + +N
Sbjct: 802 RYIPPTGHPYQG*LPVN 752
>TC89463 weakly similar to GP|13447813|gb|AAK26764.1 small basic membrane
integral protein ZmSIP1-1 {Zea mays}, partial (75%)
Length = 1142
Score = 26.6 bits (57), Expect = 6.8
Identities = 20/83 (24%), Positives = 39/83 (46%), Gaps = 3/83 (3%)
Frame = +2
Query: 115 KLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMI 174
K GDFV F + + + LGL+ T ++ F +F + + P I + + +F+I
Sbjct: 170 KAAIGDFVLTFTWMFISSTLGLVTTEIMKAFDLQFVTYNGVNY----PFIIITTLLIFII 337
Query: 175 ---FPSYRHGIGYPTSSDTNDTS 194
F + + +G + + T + S
Sbjct: 338 IFTFTAIGNALGGASFNPTGNAS 406
>TC81830 similar to GP|15982206|emb|CAC83608. Na+/H+ antiporter isoform 2
{Lycopersicon esculentum}, partial (45%)
Length = 793
Score = 26.6 bits (57), Expect = 6.8
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = +1
Query: 89 QRHYGIVTMNGLWPSP 104
+RH+GIV G WP P
Sbjct: 397 KRHFGIVDFEGQWPLP 444
>TC87279 weakly similar to GP|8843755|dbj|BAA97303.1 gene_id:MXK3.14~unknown
protein {Arabidopsis thaliana}, partial (39%)
Length = 1468
Score = 26.2 bits (56), Expect = 8.8
Identities = 15/40 (37%), Positives = 19/40 (47%), Gaps = 8/40 (20%)
Frame = -3
Query: 113 AYKLRFGDFVHAFL--------SVIVFAVLGLLDTNVVHC 144
A+ RFGDFV FL + + VLGL + HC
Sbjct: 236 AFSFRFGDFVFVFLRRSNRLGIRLGIIGVLGLSLSTFPHC 117
>TC85220 weakly similar to PIR|S33520|S33520 Lea protein - soybean, partial
(69%)
Length = 1880
Score = 26.2 bits (56), Expect = 8.8
Identities = 10/31 (32%), Positives = 17/31 (54%)
Frame = -3
Query: 55 CSTINKYLSGILLVICGFNCAFTSFTDSYTG 85
CS ++ S ++C C F+S +S+TG
Sbjct: 693 CSLLSSISSIFFGLLCSTTCVFSSLMNSFTG 601
>TC91401 similar to PIR|H96582|H96582 F15I1.27 [imported] - Arabidopsis
thaliana, partial (25%)
Length = 1377
Score = 26.2 bits (56), Expect = 8.8
Identities = 9/24 (37%), Positives = 17/24 (70%)
Frame = -2
Query: 41 LFQYLSPVVTNNGHCSTINKYLSG 64
LFQ+LS +++++GHC + + G
Sbjct: 392 LFQWLSKILSSSGHCGSSSSGTQG 321
>TC86708 similar to GP|21553407|gb|AAM62500.1 unknown {Arabidopsis thaliana},
partial (93%)
Length = 1446
Score = 26.2 bits (56), Expect = 8.8
Identities = 12/35 (34%), Positives = 20/35 (56%)
Frame = -2
Query: 38 TVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGF 72
++F+F+Y+S T HC NK+ + L +I F
Sbjct: 1421 SIFVFKYISSSTTVCSHCFLQNKFQTEPLRLILKF 1317
>BG648846 weakly similar to GP|8843755|dbj| gene_id:MXK3.14~unknown protein
{Arabidopsis thaliana}, partial (18%)
Length = 829
Score = 26.2 bits (56), Expect = 8.8
Identities = 15/40 (37%), Positives = 19/40 (47%), Gaps = 8/40 (20%)
Frame = -3
Query: 113 AYKLRFGDFVHAFL--------SVIVFAVLGLLDTNVVHC 144
A+ RFGDFV FL + + VLGL + HC
Sbjct: 206 AFSFRFGDFVFVFLRRSNRLGIRLGIIGVLGLSLSTFPHC 87
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,173,055
Number of Sequences: 36976
Number of extensions: 88429
Number of successful extensions: 458
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 455
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 457
length of query: 198
length of database: 9,014,727
effective HSP length: 91
effective length of query: 107
effective length of database: 5,649,911
effective search space: 604540477
effective search space used: 604540477
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146553.13


