

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.7 + phase: 0 /pseudo
(720 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
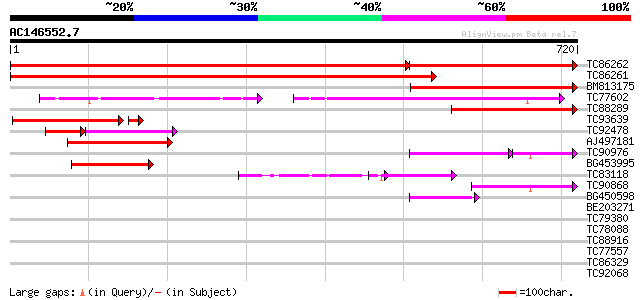
Score E
Sequences producing significant alignments: (bits) Value
TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 942 0.0
TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 1029 0.0
BM813175 similar to GP|22136936|gb putative cullin 1 protein {Ar... 373 e-103
TC77602 similar to GP|20268719|gb|AAM14063.1 putative cullin {Ar... 254 1e-91
TC88289 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 290 1e-78
TC93639 similar to GP|22335691|dbj|BAC10548. cullin-like protein... 239 9e-67
TC92478 similar to GP|22335691|dbj|BAC10548. cullin-like protein... 85 9e-35
AJ497181 weakly similar to GP|22335691|dbj cullin-like protein1 ... 137 2e-32
TC90976 similar to PIR|E96718|E96718 probable cullin T6C23.13 [i... 87 1e-27
BG453995 weakly similar to GP|14091836|gb| Putative cullin {Oryz... 106 3e-23
TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [i... 81 1e-15
TC90868 similar to GP|9295728|gb|AAF87034.1| T24P13.25 {Arabidop... 77 2e-14
BG450598 similar to GP|23429518|gb APC2 {Arabidopsis thaliana}, ... 49 5e-06
BE203271 similar to GP|22550314|gb| putative cullin protein {Ole... 35 0.077
TC79380 similar to PIR|F86392|F86392 hypothetical protein AAF985... 31 1.9
TC78088 similar to GP|11991506|emb|CAC19656. dynamin-like protei... 30 3.3
TC88916 similar to GP|20196874|gb|AAB87583.2 putative potassium ... 30 4.2
TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 29 5.5
TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase ... 29 7.2
TC92068 weakly similar to PIR|A86293|A86293 hypothetical protein... 29 7.2
>TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, complete
Length = 2332
Score = 942 bits (2436), Expect(2) = 0.0
Identities = 476/509 (93%), Positives = 479/509 (93%), Gaps = 1/509 (0%)
Frame = +1
Query: 1 MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS 60
MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS
Sbjct: 121 MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS 300
Query: 61 QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF 120
QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF
Sbjct: 301 QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF 480
Query: 121 IARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 180
IARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE
Sbjct: 481 IARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 660
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPD-YMLKAEECLRREKDRVAHY 239
IGMGKMDHYENDFEADMLKDT K Y + LRREKDRVAHY
Sbjct: 661 IGMGKMDHYENDFEADMLKDTICLLFSKGFKLDPRRFLS*LYASRLRSALRREKDRVAHY 840
Query: 240 LHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLD 299
LHSSSEPKLL KVQNELLSVYASQLLEKEHSGCHALL+DDKCEDLSRMFRLFSKIP GLD
Sbjct: 841 LHSSSEPKLLTKVQNELLSVYASQLLEKEHSGCHALLKDDKCEDLSRMFRLFSKIPSGLD 1020
Query: 300 PVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNS 359
PVSSIFKQHVTTEGMALVKHAEDAASNKKA+KRDIVGT EQVFVR VIELHDKYLAYVNS
Sbjct: 1021PVSSIFKQHVTTEGMALVKHAEDAASNKKAQKRDIVGTHEQVFVRIVIELHDKYLAYVNS 1200
Query: 360 CFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEK 419
C QNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEK
Sbjct: 1201CLQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEK 1380
Query: 420 VVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMV 479
VVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMV
Sbjct: 1381VVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMV 1560
Query: 480 TDLTLAKENQTSFEEYLSNTPNADPGIDL 508
TDLTLAKENQTSFEEYLSNTPNADPGIDL
Sbjct: 1561TDLTLAKENQTSFEEYLSNTPNADPGIDL 1647
Score = 419 bits (1077), Expect(2) = 0.0
Identities = 211/213 (99%), Positives = 211/213 (99%)
Frame = +2
Query: 508 LTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF 567
LTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF
Sbjct: 1646 LTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF 1825
Query: 568 DPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKE 627
DPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKE
Sbjct: 1826 DPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKE 2005
Query: 628 PNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKS 687
PNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKS
Sbjct: 2006 PNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKS 2185
Query: 688 RKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
RKVLGYQQLVMECVE LGRMFKPDVKAI KRIE
Sbjct: 2186 RKVLGYQQLVMECVEXLGRMFKPDVKAIKKRIE 2284
>TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (72%)
Length = 1742
Score = 1029 bits (2661), Expect = 0.0
Identities = 507/541 (93%), Positives = 523/541 (95%)
Frame = +3
Query: 1 MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS 60
MSERKTIDLEQGWDFM +GI KLKNILEGLPEPQFS EDYMMLYTTIYNMCTQKPPNDYS
Sbjct: 120 MSERKTIDLEQGWDFMLKGIQKLKNILEGLPEPQFSSEDYMMLYTTIYNMCTQKPPNDYS 299
Query: 61 QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF 120
QPLYDKY+EAFEEYI+STVLPSLREKHDEFMLRELV+RWANHKIMVRWLSRFFHYLDRYF
Sbjct: 300 QPLYDKYREAFEEYILSTVLPSLREKHDEFMLRELVRRWANHKIMVRWLSRFFHYLDRYF 479
Query: 121 IARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 180
IARRSLPPLNEVGL CFRDLVYKE++GK+RDA+ISLIDQEREGEQIDRAL+KNVLDIFVE
Sbjct: 480 IARRSLPPLNEVGLTCFRDLVYKEVNGKVRDAVISLIDQEREGEQIDRALIKNVLDIFVE 659
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYL 240
IGMG MDHYENDFE MLKDTSAYYSRKASNWILEDSCPDYMLKAEECL+REKDRVA+YL
Sbjct: 660 IGMGHMDHYENDFEVAMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLKREKDRVANYL 839
Query: 241 HSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDP 300
HSSSEPKLLEKVQ+ELLSVYA+QLLEKEHSGCHALLRDDK EDLSRMFRLFSKIP+GLDP
Sbjct: 840 HSSSEPKLLEKVQHELLSVYANQLLEKEHSGCHALLRDDKVEDLSRMFRLFSKIPKGLDP 1019
Query: 301 VSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSC 360
VSSIFKQHVT EG LVK AEDAASNKK EKRDIVG QEQVFVRKVIELHDKYLAYVN C
Sbjct: 1020VSSIFKQHVTEEGTTLVKLAEDAASNKKPEKRDIVGLQEQVFVRKVIELHDKYLAYVNDC 1199
Query: 361 FQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 420
FQNHTLFHKALKEAFE+FCNKGVAG+SSAELLATFCDNILKKGGSEKLSDEAIEETLEKV
Sbjct: 1200FQNHTLFHKALKEAFEIFCNKGVAGSSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 1379
Query: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT
Sbjct: 1380VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 1559
Query: 481 DLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 540
DLTLAKENQTSFEEYL N PN DPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK
Sbjct: 1560DLTLAKENQTSFEEYLKNNPNVDPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 1739
Query: 541 E 541
E
Sbjct: 1740E 1742
>BM813175 similar to GP|22136936|gb putative cullin 1 protein {Arabidopsis
thaliana}, partial (30%)
Length = 680
Score = 373 bits (957), Expect = e-103
Identities = 181/211 (85%), Positives = 200/211 (94%)
Frame = +1
Query: 510 VTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDP 569
VTVLTT WPSYKSFDLNLP+EMVKCVEVFK FY TKTKHRKLTWIYSLGTCNI GKF+P
Sbjct: 1 VTVLTTVLWPSYKSFDLNLPSEMVKCVEVFKGFYETKTKHRKLTWIYSLGTCNIIGKFEP 180
Query: 570 KTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPN 629
KT+EL+V+TYQA+ALLLFN++D+LSYSEIMTQLNL +ED++RLLHSLSCAKYKIL KEPN
Sbjct: 181 KTIELIVSTYQAAALLLFNTADKLSYSEIMTQLNLTNEDLVRLLHSLSCAKYKILAKEPN 360
Query: 630 TKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRK 689
T+TI P D FEFN+KFTDKMRRIKIPLPPVDE+KKVIEDVDKDRRYAIDA+IVRIMKSRK
Sbjct: 361 TRTISPNDSFEFNSKFTDKMRRIKIPLPPVDERKKVIEDVDKDRRYAIDAAIVRIMKSRK 540
Query: 690 VLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
VLG+QQLV+ECVEQLGRMFKPD+KAI KRIE
Sbjct: 541 VLGHQQLVLECVEQLGRMFKPDIKAIKKRIE 633
>TC77602 similar to GP|20268719|gb|AAM14063.1 putative cullin {Arabidopsis
thaliana}, partial (87%)
Length = 2708
Score = 254 bits (649), Expect(2) = 1e-91
Identities = 140/348 (40%), Positives = 209/348 (59%), Gaps = 4/348 (1%)
Frame = +2
Query: 361 FQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 420
F + F +K+AFE N + N AEL+A F D+ L+ G++ S+E +E TL+KV
Sbjct: 974 FAKNEAFSNTIKDAFEHLIN--LRQNRPAELIAKFLDDKLR-AGNKGTSEEELEGTLDKV 1144
Query: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
+ L +I KD+F FY+K LA+RLL KSA+ D E+S+++KLK +CG QFT+K+EGM
Sbjct: 1145 LVLFRFIQGKDVFEAFYKKDLAKRLLLGKSASIDAEKSMISKLKTECGSQFTNKLEGMFK 1324
Query: 481 DLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 540
D+ L+KE SF + GI+++V VLTTG+WP+Y D+ LP E+ ++FK
Sbjct: 1325 DIELSKEINESFRQSSQARTKLPSGIEMSVHVLTTGYWPTYPPMDVRLPHELNVYQDIFK 1504
Query: 541 EFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMT 600
EFY +K R+L W SLG C + F EL V+ +Q L+ FN +++LS+ +I
Sbjct: 1505 EFYLSKYSGRRLMWQNSLGHCVLKADFPKGKKELAVSLFQTVVLMQFNDAEKLSFQDIKD 1684
Query: 601 QLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPL---- 656
+ D+++ R L SL+C K ++L K P + + D F FN FT + RIK+
Sbjct: 1685 STGIEDKELRRTLQSLACGKVRVLQKMPKGRDVEDYDSFVFNDTFTAPLYRIKVNAIQLK 1864
Query: 657 PPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQL 704
V+E E V +DR+Y +DA+IVRIMK+RKVL + L+ E +Q+
Sbjct: 1865 ETVEENTNTTERVFQDRQYQVDAAIVRIMKTRKVLSHTLLITELFQQV 2008
Score = 101 bits (251), Expect(2) = 1e-91
Identities = 75/290 (25%), Positives = 142/290 (48%), Gaps = 7/290 (2%)
Frame = +3
Query: 39 DYMMLYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKR 98
D LY + ++C K + LY + ++ E +I + + + + D + LV+R
Sbjct: 81 DLEKLYQAVNDLCIHKMGGN----LYQRIEKECEVHISAALQSLVGQSPDLIVFLSLVER 248
Query: 99 -W---ANHKIMVRWLSRFFHYLDRYFIARR-SLPPLNEVGLACFRD--LVYKELHGKMRD 151
W + +M+R ++ F LDR ++ + ++ + ++GL FR + E+ K
Sbjct: 249 CWQDLCDQMLMIRGIALF---LDRTYVKQSPNIRSIWDMGLQIFRKHLSLSPEVQHKTVT 419
Query: 152 AIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASN 211
++ +ID ER GE +DR LL ++L +F +G+ Y FE L+ TS +Y+ +
Sbjct: 420 GLLRMIDSERLGEAVDRTLLNHLLKMFTALGI-----YAESFEKPFLECTSEFYAAEGVK 584
Query: 212 WILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSG 271
++ + PDY+ E L+ E +R YL +S++ L+ + +LL + +L+K G
Sbjct: 585 YMQQSDVPDYLKHVETRLQEEHERCLIYLDASTKKPLITTTEKQLLERHIPAILDK---G 755
Query: 272 CHALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKHAE 321
L+ ++ EDL RM LFS++ L+ + ++ G +V E
Sbjct: 756 FSMLMDGNRIEDLQRMHLLFSRV-NALESLRQAISSYIRRTGQGIVMDEE 902
>TC88289 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (24%)
Length = 1019
Score = 290 bits (742), Expect = 1e-78
Identities = 147/160 (91%), Positives = 153/160 (94%)
Frame = +2
Query: 561 CNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAK 620
CNISGKF+PKT+EL+VTTYQASALLLFNSSDRLSYSEIM QLNL D+DVIRLLHSLSCAK
Sbjct: 2 CNISGKFEPKTMELIVTTYQASALLLFNSSDRLSYSEIMAQLNLTDDDVIRLLHSLSCAK 181
Query: 621 YKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDAS 680
YKIL KEP+TK ILPTD FEFN+KFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDAS
Sbjct: 182 YKILNKEPSTKAILPTDSFEFNSKFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDAS 361
Query: 681 IVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
IVRIMKSRKVL YQQLVMECVEQLGRMFKPDVKAI KRIE
Sbjct: 362 IVRIMKSRKVLNYQQLVMECVEQLGRMFKPDVKAIKKRIE 481
>TC93639 similar to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (21%)
Length = 564
Score = 239 bits (610), Expect(2) = 9e-67
Identities = 110/141 (78%), Positives = 123/141 (87%)
Frame = +1
Query: 4 RKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPL 63
RK ID +QGW +M GI KLK ILEGLPE QF+ E+YMMLYTTIYNMCTQKPP DYSQ L
Sbjct: 61 RKVIDFDQGWAYMENGIKKLKRILEGLPETQFTSEEYMMLYTTIYNMCTQKPPLDYSQQL 240
Query: 64 YDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIAR 123
YDKYKE F+EYI STVL ++R+KHDEFMLRELV+RW NHK++VRWLSRFFHYLDRYF+AR
Sbjct: 241 YDKYKEVFDEYIRSTVLSAVRDKHDEFMLRELVQRWLNHKVLVRWLSRFFHYLDRYFVAR 420
Query: 124 RSLPPLNEVGLACFRDLVYKE 144
RSLPPLN VGL+ FRDLVY E
Sbjct: 421 RSLPPLNAVGLSAFRDLVYME 483
Score = 33.5 bits (75), Expect(2) = 9e-67
Identities = 15/19 (78%), Positives = 18/19 (93%)
Frame = +2
Query: 152 AIISLIDQEREGEQIDRAL 170
A+I LID+EREGEQIDR+L
Sbjct: 506 AVIVLIDKEREGEQIDRSL 562
>TC92478 similar to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (7%)
Length = 716
Score = 84.7 bits (208), Expect(2) = 9e-35
Identities = 46/118 (38%), Positives = 68/118 (56%), Gaps = 1/118 (0%)
Frame = +3
Query: 97 KRWANHKIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISL 156
+RW+NHKIM + LSR F YL +Y I +R L L E G F LVY E+H ++ DAI+++
Sbjct: 159 ERWSNHKIMTKLLSRIFRYLHKYHIRKRGLSSLEETGFLSFYYLVYDEMHRQVMDAILAM 338
Query: 157 IDQEREGEQIDRALLKNVLDIFV-EIGMGKMDHYENDFEADMLKDTSAYYSRKASNWI 213
ID++R GE ID+ L+ N + + K + + + + S Y +ASNWI
Sbjct: 339 IDRKRAGEPIDQTLVNNAFGFLLRDWRKHKKERPKTLCRNND*RICSILYYVEASNWI 512
Score = 81.3 bits (199), Expect(2) = 9e-35
Identities = 37/51 (72%), Positives = 44/51 (85%)
Frame = +2
Query: 46 TIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELV 96
TIY MCTQKPP++YS+ LY+KYKE F+ YI STVLPSLREK DE +LREL+
Sbjct: 5 TIYVMCTQKPPHEYSEQLYEKYKETFDGYIKSTVLPSLREKKDELLLRELL 157
>AJ497181 weakly similar to GP|22335691|dbj cullin-like protein1 {Pisum
sativum}, partial (10%)
Length = 461
Score = 137 bits (344), Expect = 2e-32
Identities = 67/133 (50%), Positives = 90/133 (67%)
Frame = +3
Query: 74 YIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIARRSLPPLNEVG 133
YIVSTVLPSLR K DE +LREL+ RW+ HK M + LS+FFHYLDRYFI R+ LP L E+G
Sbjct: 9 YIVSTVLPSLRGKKDELLLRELLGRWSIHKTMTKCLSKFFHYLDRYFIGRQRLPSLEEIG 188
Query: 134 LACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDF 193
L F DLVY E+H ++ DAI+++ID++ GE ID L+ N L + EIG + F
Sbjct: 189 LLSFYDLVYVEMHREVMDAILAMIDRKWAGEPIDETLVHNALTFYSEIGESTGKNDPKHF 368
Query: 194 EADMLKDTSAYYS 206
+K+ + +Y+
Sbjct: 369 AETTIKENATFYT 407
>TC90976 similar to PIR|E96718|E96718 probable cullin T6C23.13 [imported] -
Arabidopsis thaliana, partial (33%)
Length = 825
Score = 87.4 bits (215), Expect(2) = 1e-27
Identities = 52/134 (38%), Positives = 75/134 (55%), Gaps = 2/134 (1%)
Frame = +3
Query: 508 LTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF 567
LTV VLTTG WP+ S NLP E+ E F+ +Y R+L+W ++G ++ F
Sbjct: 6 LTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATF 185
Query: 568 DP-KTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYK-ILI 625
+ EL V+TYQ L+LFN++D+LSY EI + D+ R L SL+ K + +L
Sbjct: 186 GKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLR 365
Query: 626 KEPNTKTILPTDYF 639
KEP +K + D F
Sbjct: 366 KEPMSKDVGEDDAF 407
Score = 54.7 bits (130), Expect(2) = 1e-27
Identities = 32/87 (36%), Positives = 51/87 (57%), Gaps = 5/87 (5%)
Frame = +1
Query: 639 FEFNAKFTDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGY 693
F N KF+ K+ ++KI EK++ + V++DR+ I+A+IVRIMKSR++L +
Sbjct: 406 FSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDH 585
Query: 694 QQLVMECVEQLGRMFKPDVKAI*KRIE 720
L+ E +QL F + + KRIE
Sbjct: 586 NNLIAEVTKQLQLRFLANPTEVKKRIE 666
>BG453995 weakly similar to GP|14091836|gb| Putative cullin {Oryza sativa},
partial (6%)
Length = 670
Score = 106 bits (265), Expect = 3e-23
Identities = 55/105 (52%), Positives = 71/105 (67%), Gaps = 1/105 (0%)
Frame = +2
Query: 79 VLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIARR-SLPPLNEVGLACF 137
VLPSLRE DE +LREL+ RW+NHKIM + LSR F YLDR+ I R+ LP L E G F
Sbjct: 344 VLPSLRENKDELLLRELLIRWSNHKIMTKLLSRIFCYLDRFHIPRKWGLPSLEETGFLSF 523
Query: 138 RDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIG 182
LVY E+ ++ DAI+++ID + GE ID+ L+ N L + EIG
Sbjct: 524 YHLVYDEVKKQVMDAILAMIDWRQAGEPIDQTLVNNALAFYSEIG 658
>TC83118 similar to PIR|E96718|E96718 probable cullin T6C23.13 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 902
Score = 81.3 bits (199), Expect = 1e-15
Identities = 60/201 (29%), Positives = 98/201 (47%), Gaps = 9/201 (4%)
Frame = +2
Query: 291 FSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELH 350
F ++ GL + + H+ G LV E +D V FV+++++
Sbjct: 2 FRRVADGLLKIREVMTLHIRESGKQLVTDPE--------RLKDPVE-----FVQRLLDEK 142
Query: 351 DKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSD 410
DKY +N F N F AL +FE F N S E ++ F D+ L+KG + +++
Sbjct: 143 DKYDKIINQAFNNDKSFQNALNSSFEYFIN---LNPRSPEFISLFVDDKLRKG-LKGVNE 310
Query: 411 EAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQ--CG 468
+ +E TL+KV+ L Y+ +KD+F ++Y++ LA+ E+ L LK++ C
Sbjct: 311 DDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKEAFV-------LEKRFLMMLKERSHC* 469
Query: 469 GQ-------FTSKMEGMVTDL 482
Q FTSK+EGM TD+
Sbjct: 470 AQD*MWAINFTSKLEGMFTDM 532
Score = 59.7 bits (143), Expect = 4e-09
Identities = 33/112 (29%), Positives = 57/112 (50%)
Frame = +3
Query: 456 ERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTT 515
++ ++ KLK +CG + + L K +Q + + + ++ P+ G LTV VLTT
Sbjct: 450 KKGLIVKLKTECG--LSILHQN*RECLQT*KPSQDTMQGFYASHPDLGDGPTLTVQVLTT 623
Query: 516 GFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF 567
G WP+ S NLP E+ E F+ +Y R+L+W ++G ++ F
Sbjct: 624 GSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATF 779
>TC90868 similar to GP|9295728|gb|AAF87034.1| T24P13.25 {Arabidopsis
thaliana}, partial (22%)
Length = 779
Score = 77.0 bits (188), Expect = 2e-14
Identities = 50/140 (35%), Positives = 79/140 (55%), Gaps = 6/140 (4%)
Frame = +3
Query: 587 FNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYK-ILIKEPNTKTILPTDYFEFNAKF 645
F+++DR++ EI + D+ R L SL+ K K +L KEP +K I D F FN KF
Sbjct: 12 FHNADRMTCKEIEQATAIPMSDLKRCLQSLALVKGKNVLRKEPMSKDISEDDVFFFNEKF 191
Query: 646 TDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMEC 700
T K+ ++KI E + + V+++R+ I+ +IVR+MKSR+VL + ++ E
Sbjct: 192 TSKLFKVKIGTVVAQRETEPENIETRQRVEEERKPQIETAIVRVMKSRRVLEHNNVIAEV 371
Query: 701 VEQLGRMFKPDVKAI*KRIE 720
+QL F P+ I K+IE
Sbjct: 372 TKQLQARFLPNPVVIKKQIE 431
>BG450598 similar to GP|23429518|gb APC2 {Arabidopsis thaliana}, partial
(13%)
Length = 604
Score = 49.3 bits (116), Expect = 5e-06
Identities = 25/89 (28%), Positives = 48/89 (53%)
Frame = +2
Query: 508 LTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF 567
+ T++++ FWP + LNLP + K + + + +S RKL W SLGT + +F
Sbjct: 62 VAATIISSNFWPPIQDEPLNLPEPVDKLLSDYAKRFSEVKTPRKLQWKKSLGTVKLELQF 241
Query: 568 DPKTVELVVTTYQASALLLFNSSDRLSYS 596
+ + ++ V AS ++ F D++S++
Sbjct: 242 EDREMQFTVAPVLASIIMKF--QDQMSWT 322
>BE203271 similar to GP|22550314|gb| putative cullin protein {Olea europaea},
partial (12%)
Length = 336
Score = 35.4 bits (80), Expect = 0.077
Identities = 15/34 (44%), Positives = 21/34 (61%)
Frame = +1
Query: 534 KCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF 567
+ V++FKEFY +K R+L W SLG C + F
Sbjct: 226 EAVDIFKEFYLSKYSGRRLMWQNSLGHCVLKADF 327
>TC79380 similar to PIR|F86392|F86392 hypothetical protein AAF98564.1
[imported] - Arabidopsis thaliana, partial (49%)
Length = 978
Score = 30.8 bits (68), Expect = 1.9
Identities = 18/71 (25%), Positives = 27/71 (37%), Gaps = 11/71 (15%)
Frame = +1
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSR-----------KASNWILEDSCPDYMLKAEECL 229
I +DHY A + K Y + K W+ D D+ AE C
Sbjct: 514 INQAGIDHYNKFINALLAKGIEPYVTLYHWDLPQALDDKYKGWLSTDIIKDFATYAETCF 693
Query: 230 RREKDRVAHYL 240
++ DRV H++
Sbjct: 694 QKFGDRVKHWI 726
>TC78088 similar to GP|11991506|emb|CAC19656. dynamin-like protein DLP1
{Arabidopsis thaliana}, partial (98%)
Length = 2343
Score = 30.0 bits (66), Expect = 3.3
Identities = 14/39 (35%), Positives = 24/39 (60%), Gaps = 3/39 (7%)
Frame = +3
Query: 674 RYAIDASIVRIM---KSRKVLGYQQLVMECVEQLGRMFK 709
R+AID ++ + +RKV GY+QL+ C+ G ++K
Sbjct: 2124 RFAIDFVVLLFL*SNSNRKV*GYKQLICTCIYSFGPVYK 2240
>TC88916 similar to GP|20196874|gb|AAB87583.2 putative potassium transporter
{Arabidopsis thaliana}, partial (30%)
Length = 1162
Score = 29.6 bits (65), Expect = 4.2
Identities = 19/47 (40%), Positives = 24/47 (50%), Gaps = 4/47 (8%)
Frame = +1
Query: 488 NQTSFEEYLSNTPNAD----PGIDLTVTVLTTGFWPSYKSFDLNLPA 530
N+ S E L+ P+ PGI L T LTTG ++ F NLPA
Sbjct: 127 NKVSLEWLLALGPSLGIARVPGIGLVFTDLTTGIPANFSRFVTNLPA 267
>TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (88%)
Length = 2682
Score = 29.3 bits (64), Expect = 5.5
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +3
Query: 500 PNADPGIDLTVTVLTTGFWPSYKSFD 525
P + G ++ V VL TG WP KSF+
Sbjct: 606 PESSSGNEVVVGVLDTGVWPESKSFN 683
>TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (91%)
Length = 2671
Score = 28.9 bits (63), Expect = 7.2
Identities = 12/33 (36%), Positives = 20/33 (60%)
Frame = +2
Query: 493 EEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFD 525
E+ ++ P++ ++ V V+ TG WP KSFD
Sbjct: 437 EKTITLLPSSGKQSEVIVGVIDTGVWPELKSFD 535
>TC92068 weakly similar to PIR|A86293|A86293 hypothetical protein AAF82155.1
[imported] - Arabidopsis thaliana, partial (77%)
Length = 912
Score = 28.9 bits (63), Expect = 7.2
Identities = 27/92 (29%), Positives = 43/92 (46%), Gaps = 4/92 (4%)
Frame = +1
Query: 170 LLKNVLDIFVEIGMGKMDHYEND---FEADMLKDTSAYYSRKASNW-ILEDSCPDYMLKA 225
LL L I E MDH+ N F+ D +Y S +A W ++ D+ D++ +
Sbjct: 523 LLTEYLKIQNEYRALTMDHWRNFYRFFKEVSFTDLQSYDSSQA--WPVIVDNFVDWLKEK 696
Query: 226 EECLRREKDRVAHYLHSSSEPKLLEKVQNELL 257
E+ + V+ LHS SEP+ K+ N +
Sbjct: 697 EKTI*L*PPEVSVQLHSYSEPEYRLKLINNCI 792
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,850,987
Number of Sequences: 36976
Number of extensions: 282685
Number of successful extensions: 1482
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1457
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1477
length of query: 720
length of database: 9,014,727
effective HSP length: 103
effective length of query: 617
effective length of database: 5,206,199
effective search space: 3212224783
effective search space used: 3212224783
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146552.7


