

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.4 + phase: 0
(340 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
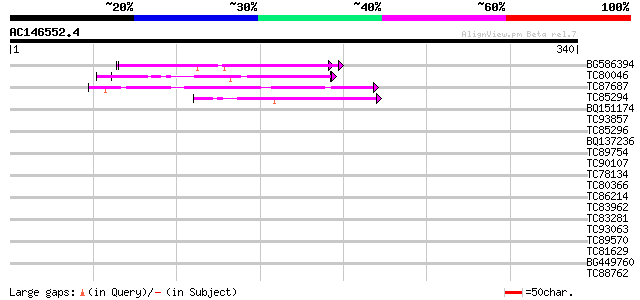
Score E
Sequences producing significant alignments: (bits) Value
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 56 2e-08
TC80046 similar to SP|P22699|GUN6_DICDI Endoglucanase precursor ... 51 7e-07
TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 42 4e-04
TC85294 similar to GP|2961298|emb|CAA12357.1 hypothetical protei... 41 8e-04
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 40 0.002
TC93857 similar to SP|Q05049|MUC1_XENLA Integumentary mucin C.1 ... 39 0.003
TC85296 weakly similar to GP|2505870|emb|CAA72908.1 hypothetical... 38 0.005
BQ137236 weakly similar to GP|19070096|emb UL49 tegument protein... 38 0.006
TC89754 similar to GP|21618273|gb|AAM67323.1 unknown {Arabidopsi... 38 0.006
TC90107 similar to PIR|F96782|F96782 hypothetical protein F22H5.... 37 0.008
TC78134 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Ar... 37 0.011
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 37 0.014
TC86214 similar to PIR|B86201|B86201 protein F12K11.9 [imported]... 36 0.024
TC83962 similar to PIR|B86289|B86289 hypothetical protein AAF719... 36 0.024
TC83281 similar to GP|3450826|gb|AAC32596.1| serum opacity facto... 35 0.032
TC93063 similar to PIR|T08852|T08852 lustrin A - California red ... 35 0.032
TC89570 similar to GP|13936308|gb|AAK40307.1 putative methyl-bin... 34 0.092
TC81629 similar to GP|15081654|gb|AAK82482.1 AT4g26750/F10M23_90... 34 0.092
BG449760 33 0.16
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 33 0.21
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 56.2 bits (134), Expect = 2e-08
Identities = 35/135 (25%), Positives = 64/135 (46%), Gaps = 5/135 (3%)
Frame = +1
Query: 65 TTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQP---TPTKASKTVP 121
TT P TT SE + T SET + +T+ + LG + TPTK+ VP
Sbjct: 13 TTTQPEKETTTTESET-NTPTKSETDVPETTTTIEKSKNDLGTTTESETETPTKSETNVP 189
Query: 122 EKTVLE--TQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDSPTIID 179
E T E QT+++ P + +++T ++ P+ T+ + + T + P++ T ++
Sbjct: 190 ETTTTEPIVQTDSEETPTEKPEKETTTKTTKPEAETTTEPETNTPTKSETDVPETTTTLE 369
Query: 180 LTSDQPSISNTTQTE 194
+ P + ++TE
Sbjct: 370 KNQNVPGSTTESETE 414
Score = 50.1 bits (118), Expect = 1e-06
Identities = 32/137 (23%), Positives = 65/137 (47%), Gaps = 2/137 (1%)
Frame = +1
Query: 66 TNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQP-TPTKASKTVPEKT 124
TN P +TT+ + S+ T +E P+ + + E E + TPTK+ VPE T
Sbjct: 178 TNVPETTTTEPIVQTDSEETPTEKPEKETTTKTTKPEAETTTEPETNTPTKSETDVPETT 357
Query: 125 VLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQ-TTEQQQQPDSPTIIDLTSD 183
T + Q +P T + +T + + +TT+ + P+++ T+ + + T+
Sbjct: 358 T--TLEKNQNVPGSTTESETETPTKSETETTTTEETPTEKPETDIPKTTTASEKNKTTTT 531
Query: 184 QPSISNTTQTEPSPIPD 200
+P++ ++ P+ P+
Sbjct: 532 EPTVPTVSEETPTEKPE 582
>TC80046 similar to SP|P22699|GUN6_DICDI Endoglucanase precursor (EC
3.2.1.4) (Endo-1 4-beta-glucanase) (Spore germination
protein 270-6), partial (11%)
Length = 643
Score = 50.8 bits (120), Expect = 7e-07
Identities = 42/146 (28%), Positives = 67/146 (45%), Gaps = 2/146 (1%)
Frame = +3
Query: 53 VSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPT 112
VS + + E +T +P ++T+ +E +++ T++ETP E ST T
Sbjct: 261 VSPSPTETSTETSTETPTETSTETPTETSTE-TSTETPT--ETST-------------ET 392
Query: 113 PTKASKTVPEKTVLETQTE--TQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQ 170
PT+ S P +T ET TE T+T P T +T++E P TS + P T +
Sbjct: 393 PTETSTETPTETSTETSTETSTETPPPST---ETSTETTPPSTETSTETTPPSTETSTET 563
Query: 171 QPDSPTIIDLTSDQPSISNTTQTEPS 196
P S T+ + ++T T PS
Sbjct: 564 TPPSTETSTETTPPSTETSTENTPPS 641
Score = 47.4 bits (111), Expect = 8e-06
Identities = 37/140 (26%), Positives = 61/140 (43%), Gaps = 6/140 (4%)
Frame = +3
Query: 62 PEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVP 121
P T++ +TT V +T + +P E ST + E TPT+ S P
Sbjct: 183 PVTVTSTATITTTVSPITVTDTSTNTVSPSPTETSTETSTE---------TPTETSTETP 335
Query: 122 EKTVLETQTE------TQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDSP 175
+T ET TE T+T E + + T + +T+++ PS +T+ + P +
Sbjct: 336 TETSTETSTETPTETSTETPTETSTETPTETSTETSTETSTETPPPSTETSTETTPPSTE 515
Query: 176 TIIDLTSDQPSISNTTQTEP 195
T + T PS +T+T P
Sbjct: 516 TSTETT--PPSTETSTETTP 569
>TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (67%)
Length = 1969
Score = 41.6 bits (96), Expect = 4e-04
Identities = 37/177 (20%), Positives = 77/177 (42%), Gaps = 3/177 (1%)
Frame = +3
Query: 48 DNPEPVSVQ--QPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHL 105
++P P + QP+PNPEP N + + T++ S+ + D E+ +E ST +
Sbjct: 84 ESPVPADKESDQPKPNPEPVEN--NTTETEEVSKPSGDDKIPESGSFKEEST-------I 236
Query: 106 GGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQT 165
GE+ KA + + + P T ++ P P+++T
Sbjct: 237 VGELPEAEKKALAELKQLIQEALNKHEFSAPASTTPSPVKEQKPEP-----TPEAPAEET 401
Query: 166 TEQQQQ-PDSPTIIDLTSDQPSISNTTQTEPSPIPDHILESEYIEEQLIRLSDEIQA 221
++ +Q ++ +++ +T+ +S T P P + E+E +E++ + E+ A
Sbjct: 402 NKKDEQVSETESVVAVTTADDDVST---TPPPPPTEAEAEAEQPKEEVEKKETEVAA 563
Score = 36.6 bits (83), Expect = 0.014
Identities = 32/133 (24%), Positives = 60/133 (45%), Gaps = 5/133 (3%)
Frame = +3
Query: 50 PEPVSVQQPQPNPEPTTNSPHNSTTQKASEVA-SDATTSETPQHQEYSTLHNLEKHLGGE 108
P PV Q+P+P PE +P T +K +V+ +++ + T + ST
Sbjct: 342 PSPVKEQKPEPTPE----APAEETNKKDEQVSETESVVAVTTADDDVSTT--------PP 485
Query: 109 MQPTPTKASKTVPEKTV--LETQTETQTIPEQTVQEQTASEQ--VAPDQTTSDQHIPSDQ 164
PT +A P++ V ET+ ++ E + A E+ VA + ++ +
Sbjct: 486 PPPTEAEAEAEQPKEEVEKKETEVAASSVDEDGAKTVEAIEESVVAVASSVPEEPKVVEA 665
Query: 165 TTEQQQQPDSPTI 177
++ +QQQP+ +I
Sbjct: 666 SSPEQQQPEEVSI 704
>TC85294 similar to GP|2961298|emb|CAA12357.1 hypothetical protein {Cicer
arietinum}, partial (61%)
Length = 1353
Score = 40.8 bits (94), Expect = 8e-04
Identities = 31/117 (26%), Positives = 55/117 (46%), Gaps = 4/117 (3%)
Frame = +2
Query: 111 PTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSD---QHIPSDQTTE 167
P PT +T PE V++T E+T+ EQTA E AP+Q ++ +P+++TTE
Sbjct: 293 PPPTTVPETTPE--VIKT--------EETIPEQTAIEVPAPEQPEAEVPATEVPTEETTE 442
Query: 168 QQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPDHILES-EYIEEQLIRLSDEIQALI 223
Q + T + + P T + + E+ + EE + + +E A++
Sbjct: 443 QPTETTEETPAAVETQDPVEVETKEVVTEEAKEENTEAPKETEESVEEVKEEAAAVV 613
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 39.7 bits (91), Expect = 0.002
Identities = 33/134 (24%), Positives = 47/134 (34%)
Frame = +1
Query: 36 KFNLPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEY 95
K N PP P P +P P PQP P+P PH+ + E T Q
Sbjct: 208 KNNTPPPPPHTPPDPTP-----PQPPPQP-PKPPHHEKRPRTPEPPGGRPPPPTTPKQAR 369
Query: 96 STLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTT 155
T G +P P T E T +T+ Q P+Q +E+ P +
Sbjct: 370 PTTQERGGARGARPRPPPAPPQGTGGEDT--KTRPGKQREPQQNPRERGTGHARTPGKKN 543
Query: 156 SDQHIPSDQTTEQQ 169
+ H T ++
Sbjct: 544 TPPHTKKKGETHKK 585
Score = 28.1 bits (61), Expect = 5.1
Identities = 32/145 (22%), Positives = 48/145 (33%), Gaps = 3/145 (2%)
Frame = +3
Query: 58 PQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKAS 117
P P+P+ T + + QK S T ++P+ +E K P P A
Sbjct: 15 PTPSPQATPQRKGHPS-QKPPTGRSHKKTKKSPRKKE--------KPPPRPATPPPAPAP 167
Query: 118 KTVPEKTVLETQTETQ---TIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDS 174
P +T T + Q T P + T QTT + P+D T + P +
Sbjct: 168 PGPPNQTTNTTNPKKQHTPTTPTHPPRPHTPPTPPPTTQTTPPREAPTDPGTTRGTPPTA 347
Query: 175 PTIIDLTSDQPSISNTTQTEPSPIP 199
+ P P P P
Sbjct: 348 DDAQTSPAHHPGEGGGKGGAPPPPP 422
>TC93857 similar to SP|Q05049|MUC1_XENLA Integumentary mucin C.1 (FIM-C.1)
(Fragment). [African clawed frog] {Xenopus laevis},
partial (8%)
Length = 763
Score = 38.9 bits (89), Expect = 0.003
Identities = 29/137 (21%), Positives = 51/137 (37%), Gaps = 2/137 (1%)
Frame = +2
Query: 41 PTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHN 100
P+H + N Q+P+PNP TT + + ++ T P H +T +
Sbjct: 176 PSHTITTTNV----TQKPKPNPTTTTTNVTPKPKPNPTTTTTNVTPKPKPNHTT-TTTNV 340
Query: 101 LEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHI 160
K T K P T T + P T +++ P+ TT+ ++
Sbjct: 341 TPKPKPSHTTTTTNVTQKPKPNPTTTTTNVAPKPKPNHTTTTTNVTQKPKPNHTTTTTNV 520
Query: 161 PS--DQTTEQQQQPDSP 175
+ TT + P++P
Sbjct: 521 TQKPNHTTTTTKFPNNP 571
>TC85296 weakly similar to GP|2505870|emb|CAA72908.1 hypothetical protein
{Arabidopsis thaliana}, partial (3%)
Length = 774
Score = 38.1 bits (87), Expect = 0.005
Identities = 24/69 (34%), Positives = 37/69 (52%), Gaps = 3/69 (4%)
Frame = +1
Query: 111 PTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSD---QHIPSDQTTE 167
P PT +T PE V++T E+T+ EQTA E AP+Q ++ +P+++TTE
Sbjct: 586 PPPTTVPETTPE--VIKT--------EETIPEQTAIEVPAPEQPEAEVPATEVPTEETTE 735
Query: 168 QQQQPDSPT 176
Q + T
Sbjct: 736 QPTETTEET 762
>BQ137236 weakly similar to GP|19070096|emb UL49 tegument protein
{Pseudorabies virus}, partial (10%)
Length = 1224
Score = 37.7 bits (86), Expect = 0.006
Identities = 49/201 (24%), Positives = 76/201 (37%), Gaps = 19/201 (9%)
Frame = +1
Query: 50 PEPVSVQQPQPNPEPTTNSP-HNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGE 108
P P + ++P+ NP P T++ H T + ATT + Q HN GG
Sbjct: 232 PPPNTTRRPKKNPPPPTHTQRHQKETGEGERKKKTATTGPEGRKQ-----HN---RPGGR 387
Query: 109 MQPTPTKAS----KTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPD--QTTSDQHIPS 162
QP P S K E + ET +T Q ++ A D TT+ Q P
Sbjct: 388 RQPRPRTKSEGERKRKEGAKEEERERETNKKGSETKGRQPTTDTHADDTQHTTTTQRSPG 567
Query: 163 DQTTEQQQQPDSPTI-----IDLTSDQPSISNTTQTEPS-------PIPDHILESEYIEE 210
+ + Q++ + + T +Q ++ T +T + ES++ E
Sbjct: 568 PRHADGQRERHAEAAARRARVASTQEQNAVEQTQRTSATRPASARRTTKQESEESKHEET 747
Query: 211 QLIRLSDEIQALILRRTVPAP 231
+ R DE Q RR AP
Sbjct: 748 EQQRRRDETQHETERR*NNAP 810
>TC89754 similar to GP|21618273|gb|AAM67323.1 unknown {Arabidopsis
thaliana}, partial (88%)
Length = 759
Score = 37.7 bits (86), Expect = 0.006
Identities = 23/72 (31%), Positives = 33/72 (44%), Gaps = 20/72 (27%)
Frame = +2
Query: 36 KFNLPPTHPLYP-------------DNPEP-------VSVQQPQPNPEPTTNSPHNSTTQ 75
+FN PP P++P +P P VS P P+P PT++SP +S+
Sbjct: 77 RFNKPPPSPIHPPPHQSIPSSTSSASSPSPSSAHHASVSNSHPSPSPPPTSSSPSSSSPT 256
Query: 76 KASEVASDATTS 87
S AS T+S
Sbjct: 257 SWSSPASSTTSS 292
>TC90107 similar to PIR|F96782|F96782 hypothetical protein F22H5.5
[imported] - Arabidopsis thaliana, partial (34%)
Length = 973
Score = 37.4 bits (85), Expect = 0.008
Identities = 26/84 (30%), Positives = 36/84 (41%)
Frame = +3
Query: 56 QQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTK 115
QQPQP P+ ++ S T A D+ + P E S + K +M P K
Sbjct: 291 QQPQPQPQGVSSDNTISAT-----TAVDSVQTIIPVESELSNVPPHTKAPATKMPLRPRK 455
Query: 116 ASKTVPEKTVLETQTETQTIPEQT 139
K P+ T E+Q+ET P T
Sbjct: 456 IRKVSPDPTTSESQSETLKPPNST 527
>TC78134 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 1133
Score = 37.0 bits (84), Expect = 0.011
Identities = 35/145 (24%), Positives = 59/145 (40%), Gaps = 3/145 (2%)
Frame = +1
Query: 41 PTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASE---VASDATTSETPQHQEYST 97
PT PL P ++ + NP PTT ++TT K S TTS TP + +T
Sbjct: 250 PTPPLTPK-----TIPRSDSNPYPTTFVQADTTTFKQVVQMLTGSSETTSTTPTTTKPTT 414
Query: 98 LHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSD 157
N ++ L + + KT P+K + ++ + T + + S
Sbjct: 415 KPNQQQDLPQQTRNFNIPPIKTAPKKQGFKLYERRNSLKNSVLMLNTLMPNFSQNSNFSP 594
Query: 158 QHIPSDQTTEQQQQPDSPTIIDLTS 182
+ +QQQ+ SP+++D S
Sbjct: 595 R--------KQQQEILSPSLLDFPS 645
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 36.6 bits (83), Expect = 0.014
Identities = 28/97 (28%), Positives = 42/97 (42%)
Frame = +3
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
PP P P NP PV+ P P+ T +SP STT A ++ + + +P T
Sbjct: 33 PP--PSTPANPPPVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSP-----PTTP 191
Query: 100 NLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIP 136
+ GG P+P S+T P + ++T P
Sbjct: 192 AISPPSGGGTTPSP--PSRTPPSSDDSPSPPSSKTPP 296
Score = 28.1 bits (61), Expect = 5.1
Identities = 22/89 (24%), Positives = 34/89 (37%), Gaps = 7/89 (7%)
Frame = +3
Query: 38 NLPPTHPLYPDNPEPVSVQQPQPN-----PEPTT--NSPHNSTTQKASEVASDATTSETP 90
N PP P P P + P P+ P P+T NSP + T A S T+ +P
Sbjct: 54 NPPPVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSPPTTPAISPPSGGGTTPSP 233
Query: 91 QHQEYSTLHNLEKHLGGEMQPTPTKASKT 119
+ + + + P P+ S +
Sbjct: 234 PSRTPPSSDDSPSPPSSKTPPPPSPPSSS 320
>TC86214 similar to PIR|B86201|B86201 protein F12K11.9 [imported] -
Arabidopsis thaliana, partial (84%)
Length = 1763
Score = 35.8 bits (81), Expect = 0.024
Identities = 43/152 (28%), Positives = 61/152 (39%), Gaps = 4/152 (2%)
Frame = +2
Query: 41 PTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHN 100
P L +P P S Q+P+P P+P +N AS +S+ T ++T ST +
Sbjct: 128 PRQTLKQQSPWP-SKQKPKPKPKPGSN-------*LASRTSSEPTQNQTGSM*NASTTSS 283
Query: 101 LEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHI 160
G Q TP AS T E + + T P +T S A ++S HI
Sbjct: 284 ----SGAPTQQTPHAASLTDSECRL----SLNPTSPPET*LTLLISSAPATSTSSSPPHI 439
Query: 161 P----SDQTTEQQQQPDSPTIIDLTSDQPSIS 188
P S + Q P SP + S P+ S
Sbjct: 440 PLPFLSPHLPQPPQSPLSPLPLASPSPLPTAS 535
Score = 33.5 bits (75), Expect = 0.12
Identities = 25/73 (34%), Positives = 32/73 (43%)
Frame = +1
Query: 62 PEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVP 121
P T NS H +T +S TS PQH+ +TL N + T T T+
Sbjct: 4 PRNTQNSNHAATPLLSSNHIPPRFTS--PQHKNTNTLTNSNNN-------TKTNTKTTIS 156
Query: 122 EKTVLETQTETQT 134
ETQT+TQT
Sbjct: 157 MAIETETQTQTQT 195
>TC83962 similar to PIR|B86289|B86289 hypothetical protein AAF71991.1
[imported] - Arabidopsis thaliana, partial (11%)
Length = 679
Score = 35.8 bits (81), Expect = 0.024
Identities = 39/158 (24%), Positives = 60/158 (37%), Gaps = 1/158 (0%)
Frame = +2
Query: 47 PDNPEPVSVQQPQ-PNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHL 105
P N P + P+P +T++ ST+ +S + +T+S TP ++L +
Sbjct: 44 PANKSPA*ISNSSTPSPSSSTST---STSSSSSPPPTPSTSSTTPSPPPPTSLSS----- 199
Query: 106 GGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQT 165
PTPT + T T T T T T S P QT+ +PS
Sbjct: 200 -----PTPTSNHLSSA*PT---TPTNTLTAVPSTPLNSPCSNLHLPQQTSLPSPLPSSSP 355
Query: 166 TEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPDHIL 203
+ Q P S+S ++ P P+P H L
Sbjct: 356 SYQTTPPALA----------SVSLSSSATPLPLPAHSL 439
>TC83281 similar to GP|3450826|gb|AAC32596.1| serum opacity factor
{Streptococcus pyogenes}, partial (1%)
Length = 902
Score = 35.4 bits (80), Expect = 0.032
Identities = 41/181 (22%), Positives = 78/181 (42%), Gaps = 8/181 (4%)
Frame = +2
Query: 51 EPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTL--HNLEKHLGGE 108
+P + QP + P + T + E ++ ++ T+ HNL+++L +
Sbjct: 371 KPHIIDQPFEEKDENIFEPVHQQTPERKEDKQVGEPFQSDIIDKFHTIKPHNLKENLDDK 550
Query: 109 MQPTPTKASKTVPEKTVL-ETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTE 167
++ E +L + E Q +P+ + +E+ S + QH S+Q +
Sbjct: 551 VRN----------EALILHDPHKEIQQVPDTSHREEERSAE------QQHQHSTSEQPQQ 682
Query: 168 QQQQPDSPTIIDLTSDQPSISNTTQTEP-SPIPDHILESEYIEEQ----LIRLSDEIQAL 222
QQP PT I++TS++ + +N + P SP H +E +Q L + D+ A
Sbjct: 683 PPQQPLQPT-INITSNEDNNTNKNENAPTSPDMAHPRGNEQSTKQGGQKLSKTKDKFNAT 859
Query: 223 I 223
I
Sbjct: 860 I 862
>TC93063 similar to PIR|T08852|T08852 lustrin A - California red abalone,
partial (4%)
Length = 1058
Score = 35.4 bits (80), Expect = 0.032
Identities = 44/196 (22%), Positives = 76/196 (38%), Gaps = 8/196 (4%)
Frame = -1
Query: 29 NRIK----LCEKFNLPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDA 84
NR+K +CE+ +L +PL V Q P+ + + + A+E +DA
Sbjct: 770 NRVKPTCDMCEEGDLQCQYPL-------VKKQAPRKS----------NVSIGAAEDEADA 642
Query: 85 TTSETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQT 144
+ P+ + E E +P + PE E + E + PE +EQ
Sbjct: 641 EADDEPEPEP-------EPEPEAEPEPEAEPEPEAEPEPEA-EPEPEAEPEPEDEEEEQE 486
Query: 145 ASEQVAPDQTT----SDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPD 200
+ D T+ S+ P+ +T Q +P+ DLT QPS + + PD
Sbjct: 485 PDDIETIDYTSNMPVSNMLTPAVETANQDYFNSAPS--DLTYPQPSAHDIAMSAVGSYPD 312
Query: 201 HILESEYIEEQLIRLS 216
+Y ++ + S
Sbjct: 311 PAATVDYSDQHAVTTS 264
>TC89570 similar to GP|13936308|gb|AAK40307.1 putative methyl-binding domain
protein MBD105 {Zea mays}, partial (9%)
Length = 728
Score = 33.9 bits (76), Expect = 0.092
Identities = 32/136 (23%), Positives = 55/136 (39%), Gaps = 13/136 (9%)
Frame = +3
Query: 51 EPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQ 110
EPVS + P P+ P S T K +E+ + T E E T LEK+L
Sbjct: 132 EPVSFELPAPSGWTKKFFPKKSGTPKKNEIVFTSPTGE-----EIRTKRQLEKYLKTNGG 296
Query: 111 P-----------TPTKASKTVPEKTVLETQTETQTIP--EQTVQEQTASEQVAPDQTTSD 157
P TP ++S+ V EK E +T P +++ + + ++ + D+
Sbjct: 297 PNISEFDWGTGETPRRSSRIV-EKVKAAPPAEPKTDPPKKRSRKSSASKKEASEDEAEET 473
Query: 158 QHIPSDQTTEQQQQPD 173
+ + + E + D
Sbjct: 474 KDVEMQEAGETKDDKD 521
>TC81629 similar to GP|15081654|gb|AAK82482.1 AT4g26750/F10M23_90
{Arabidopsis thaliana}, partial (19%)
Length = 824
Score = 33.9 bits (76), Expect = 0.092
Identities = 42/174 (24%), Positives = 65/174 (37%), Gaps = 1/174 (0%)
Frame = +2
Query: 35 EKFNLPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQE 94
+ ++ PP YP P P P P + P + + SD++ SE HQ+
Sbjct: 56 QDYHPPPPSQDYP--PPPSQDYHPPPPSQDYQPPPPSQDYHQPPPARSDSSYSEPYNHQQ 229
Query: 95 YSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQT 154
YS + ++LG T ++P + TE+ ++P V + PD +
Sbjct: 230 YSP--DQSQNLGPNYPSHETPPPYSLPHFQSYPSFTES-SLPSVPVNH---TYYQGPDAS 391
Query: 155 TSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSP-IPDHILESEY 207
S Q P TT T + S N T EP P P + +S Y
Sbjct: 392 YSSQSAP--LTTNHSLN---------TQNSSSSRNGTVPEPKPTTPTYQYDSSY 520
>BG449760
Length = 656
Score = 33.1 bits (74), Expect = 0.16
Identities = 25/101 (24%), Positives = 48/101 (46%), Gaps = 10/101 (9%)
Frame = +2
Query: 122 EKTVLETQT--ETQTIPEQTVQEQTASEQVA--------PDQTTSDQHIPSDQTTEQQQQ 171
+KT++E +T + I E Q++++S QV TTS Q P+ + + ++
Sbjct: 329 DKTIMEMKTIDHSDKINETFTQKRSSSFQVQIPSFDSPPNSPTTSSQQYPTLNSIQNERV 508
Query: 172 PDSPTIIDLTSDQPSISNTTQTEPSPIPDHILESEYIEEQL 212
SPT ++ + S T ++ + DHI+ +E+L
Sbjct: 509 SVSPTELENKNHSTKTSKTKRSRANNGEDHIMAXRKRKEKL 631
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 32.7 bits (73), Expect = 0.21
Identities = 32/122 (26%), Positives = 46/122 (37%), Gaps = 11/122 (9%)
Frame = +3
Query: 36 KFNLPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASD---ATTS---ET 89
+FN P + P PVS P P ++ +T+ AS AS TTS T
Sbjct: 72 QFNNPTSPSSSPSASSPVSTTTSPPASTPASSPVSTTTSPPASTPASSPVPTTTSPPAPT 251
Query: 90 PQHQEYSTLHNLEKHLG-----GEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQT 144
P ST G P+ T A P T T++ +T P +++ +
Sbjct: 252 PASSPVSTNSPTASPAGSLPAAATPSPSSTTAGSPSPSSTPPGTRSSNETTPGRSIGVAS 431
Query: 145 AS 146
AS
Sbjct: 432 AS 437
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,338,500
Number of Sequences: 36976
Number of extensions: 151238
Number of successful extensions: 2169
Number of sequences better than 10.0: 165
Number of HSP's better than 10.0 without gapping: 1473
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2009
length of query: 340
length of database: 9,014,727
effective HSP length: 97
effective length of query: 243
effective length of database: 5,428,055
effective search space: 1319017365
effective search space used: 1319017365
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146552.4


