

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.2 - phase: 0 /pseudo
(392 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
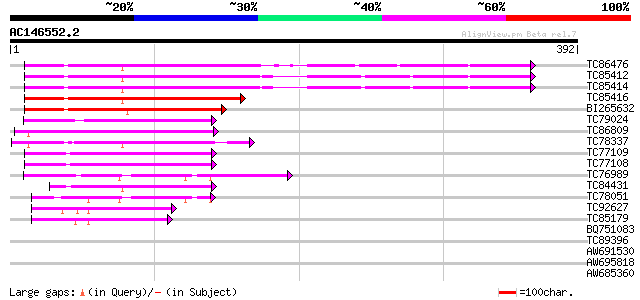
Score E
Sequences producing significant alignments: (bits) Value
TC86476 similar to GP|15450763|gb|AAK96653.1 elongation factor E... 232 2e-61
TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 225 3e-59
TC85414 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 223 8e-59
TC85416 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 147 5e-36
BI265632 homologue to GP|22947038|gb CG2238-PA {Drosophila melan... 130 1e-30
TC79024 similar to PIR|A84716|A84716 probable GTP-binding protei... 105 3e-23
TC86809 homologue to PIR|S35701|S35701 translation elongation fa... 100 1e-21
TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein... 100 2e-21
TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 97 8e-21
TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 97 8e-21
TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu ch... 71 6e-13
TC84431 similar to GP|10176983|dbj|BAB10215. GTP-binding membran... 69 2e-12
TC78051 homologue to PIR|T01400|T01400 translation elongation fa... 59 4e-09
TC92627 similar to PIR|T49975|T49975 hypothetical protein F12B17... 55 6e-08
TC85179 similar to GP|2662343|dbj|BAA23658.1 EF-1 alpha {Oryza s... 47 2e-05
BQ751083 homologue to GP|13925370|gb elongation factor 2 {Neuros... 41 0.001
TC89396 similar to SP|P48599|IFE1_ORYSA Eukaryotic translation i... 32 0.32
AW691530 similar to PIR|T01917|T01 translation initiation factor... 30 1.6
AW695818 similar to PIR|T01917|T01 translation initiation factor... 30 1.6
AW685360 homologue to PIR|T00956|T00 translation initiation fact... 29 2.7
>TC86476 similar to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 2974
Score = 232 bits (591), Expect = 2e-61
Identities = 138/369 (37%), Positives = 204/369 (54%), Gaps = 16/369 (4%)
Frame = +2
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
+ IRN+ ++AHVDHGK+TL D L+AAA G++ +VAG VR D +E R IT+KS+
Sbjct: 290 KHNIRNMSVIAHVDHGKSTLTDSLVAAA--GIIAQEVAGDVRMTDTRQDEAERGITIKST 463
Query: 71 SISLHY----------------NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVE 114
ISL+Y N + +NLIDSPGH+DF EV+ A R++DGAL++VD VE
Sbjct: 464 GISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVE 643
Query: 115 GVHIQTHAVLRQCWTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYN 174
GV +QT VLRQ E ++P L +NKMDR EL+L EAY+ + R++ VN + + Y
Sbjct: 644 GVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYE 823
Query: 175 SEKYLSDVDALLAGGTAAGGEVMEDYDDVEDKFQPQKGNVVFACALDGWGFGIHEFAEIY 234
DALL G+V + P+KG V F+ L GW F + FA++Y
Sbjct: 824 --------DALL-------GDV---------QVYPEKGTVSFSAGLHGWSFTLTNFAKMY 931
Query: 235 ASKLGGSASVGALLRALWGPWYYNPKTKMIVGKKGISGSKARPMFVQFVLEPLWQVYQGA 294
ASK G ++ LWG +++ TK K S + FVQF EP+ Q+ +
Sbjct: 932 ASKFG--VDEEKMMNRLWGENFFDSSTKKWTNKH-TSTPTCKRGFVQFCYEPIKQIIELC 1102
Query: 295 LGGGKGMVEKVIKSFNLQIQARELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVE 354
+ K + +++ + +++ E + K +++ VM WLP S A+L M++ LP P +
Sbjct: 1103MNDQKDKLWPMLQKLGVNLKSEE-KELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTK 1279
Query: 355 GQKSRISRL 363
QK R+ L
Sbjct: 1280AQKYRVENL 1306
>TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 3229
Score = 225 bits (573), Expect = 3e-59
Identities = 137/369 (37%), Positives = 197/369 (53%), Gaps = 16/369 (4%)
Frame = +3
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
+ IRN+ ++AHVDHGK+TL D L+AAA G++ +VAG VR D +E R IT+KS+
Sbjct: 213 KHNIRNMSVIAHVDHGKSTLTDSLVAAA--GIIAQEVAGDVRMTDTRADEAERGITIKST 386
Query: 71 SISLHY----------------NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVE 114
ISL+Y N + +NLIDSPGH+DF EV+ A R++DGAL++VD VE
Sbjct: 387 GISLYYEMTDESLKRFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVE 566
Query: 115 GVHIQTHAVLRQCWTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYN 174
GV +QT VLRQ E + P L +NKMDR EL + EAY R++ N I + Y
Sbjct: 567 GVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY- 743
Query: 175 SEKYLSDVDALLAGGTAAGGEVMEDYDDVEDKFQPQKGNVVFACALDGWGFGIHEFAEIY 234
+ L DV + P+KG V F+ L GW F + FA++Y
Sbjct: 744 EDPLLGDV-----------------------QVYPEKGTVAFSAGLHGWAFTLTNFAKMY 854
Query: 235 ASKLGGSASVGALLRALWGPWYYNPKTKMIVGKKGISGSKARPMFVQFVLEPLWQVYQGA 294
ASK G S ++ LWG +++P TK K S S R FVQF EP+ Q+
Sbjct: 855 ASKFGVDES--KMMERLWGENFFDPATKKWTTKNTGSASCKRG-FVQFCYEPIKQIINTC 1025
Query: 295 LGGGKGMVEKVIKSFNLQIQARELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVE 354
+ K + ++ + +++ E ++ K +++ VM WLP S A+L M++ LP P
Sbjct: 1026MNDQKDKLWPMLTKLGVTMKSEE-KDLMGKPLMKRVMQTWLPASTALLEMMIFHLPSPST 1202
Query: 355 GQKSRISRL 363
Q+ R+ L
Sbjct: 1203AQRYRVENL 1229
>TC85414 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 2892
Score = 223 bits (569), Expect = 8e-59
Identities = 136/369 (36%), Positives = 197/369 (52%), Gaps = 16/369 (4%)
Frame = +3
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
+ IRN+ ++AHVDHGK+TL D L+AAA G++ +VAG VR D +E R IT+KS+
Sbjct: 171 KHNIRNMSVIAHVDHGKSTLTDSLVAAA--GIIAQEVAGDVRMTDTRADEAERGITIKST 344
Query: 71 SISLHY----------------NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVE 114
ISL+Y N + +NLIDSPGH+DF EV+ A R++DGAL++VD VE
Sbjct: 345 GISLYYEMTDDSLKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVE 524
Query: 115 GVHIQTHAVLRQCWTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYN 174
GV +QT VLRQ E + P L +NKMDR EL + EAY R++ N I + Y
Sbjct: 525 GVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY- 701
Query: 175 SEKYLSDVDALLAGGTAAGGEVMEDYDDVEDKFQPQKGNVVFACALDGWGFGIHEFAEIY 234
+ L DV + P+KG V F+ L GW F + FA++Y
Sbjct: 702 EDPLLGDV-----------------------QVYPEKGTVAFSAGLHGWAFTLTNFAKMY 812
Query: 235 ASKLGGSASVGALLRALWGPWYYNPKTKMIVGKKGISGSKARPMFVQFVLEPLWQVYQGA 294
ASK G + ++ LWG +++P TK K S + R FVQF EP+ QV
Sbjct: 813 ASKFGVDET--KMMERLWGENFFDPATKKWTTKNTGSATCKRG-FVQFCYEPIKQVINTC 983
Query: 295 LGGGKGMVEKVIKSFNLQIQARELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVE 354
+ K + ++ + +++ E ++ K +++ VM WLP S A+L M++ LP P
Sbjct: 984 MNDQKDKLWPMLTKLGITMKSEE-KDLMGKPLMKRVMQTWLPASTALLEMMIFHLPSPST 1160
Query: 355 GQKSRISRL 363
Q+ R+ L
Sbjct: 1161AQRYRVENL 1187
>TC85416 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, partial (21%)
Length = 660
Score = 147 bits (372), Expect = 5e-36
Identities = 79/169 (46%), Positives = 106/169 (61%), Gaps = 16/169 (9%)
Frame = +3
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
+ IRN+ ++AHVDHGK+TL D L+AAA G++ +VAG VR D +E R IT+KS+
Sbjct: 156 KHNIRNMSVIAHVDHGKSTLTDSLVAAA--GIIAQEVAGDVRMTDTRADEAERGITIKST 329
Query: 71 SISLHY----------------NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVE 114
ISL+Y N + +NLIDSPGH+DF EV+ A R++DGAL++VD VE
Sbjct: 330 GISLYYEMSDESLKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVE 509
Query: 115 GVHIQTHAVLRQCWTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIV 163
GV +QT VLRQ E + P L +NKMDR EL + EAY R++
Sbjct: 510 GVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVI 656
>BI265632 homologue to GP|22947038|gb CG2238-PA {Drosophila melanogaster},
partial (21%)
Length = 593
Score = 130 bits (326), Expect = 1e-30
Identities = 70/160 (43%), Positives = 99/160 (61%), Gaps = 20/160 (12%)
Frame = +3
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
++ IRN+ ++AHVDHGK+TL D L++ A G++ AG+ RF D +EQ R IT+KS+
Sbjct: 102 KRNIRNMSVIAHVDHGKSTLTDSLVSKA--GIIAGARAGETRFTDTRKDEQDRCITIKST 275
Query: 71 SISLHYNHHT--------------------VNLIDSPGHIDFCGEVSTAARLSDGALLLV 110
+IS+ + +NLIDSPGH+DF EV+ A R++DGAL++V
Sbjct: 276 AISMFFELEEKDLVFITNPDQREKSEKGFLINLIDSPGHVDFSSEVTAALRVTDGALVVV 455
Query: 111 DAVEGVHIQTHAVLRQCWTEMLEPCLVLNKMDRLITELNL 150
D V GV +QT VLRQ E ++P L +NKMDR + EL L
Sbjct: 456 DCVSGVCVQTETVLRQAIAERIKPXLFMNKMDRALLELQL 575
>TC79024 similar to PIR|A84716|A84716 probable GTP-binding protein
[imported] - Arabidopsis thaliana, partial (67%)
Length = 1513
Score = 105 bits (262), Expect = 3e-23
Identities = 55/134 (41%), Positives = 80/134 (59%)
Frame = +2
Query: 10 DRKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKS 69
D ++RN+ ++AHVDHGKTTL D+L+ + H R MD + E+ R IT+ S
Sbjct: 191 DPNRLRNVAVIAHVDHGKTTLMDRLLRQCGADIPHE------RAMDSISLERERGITISS 352
Query: 70 SSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWT 129
S+ + + +N++D+PGH DF GEV + +GA+L+VDA EG QT VL +
Sbjct: 353 KVTSISWKDNELNMVDTPGHADFGGEVERVVGMVEGAVLVVDAGEGPLAQTKFVLAKALK 532
Query: 130 EMLEPCLVLNKMDR 143
L P L+LNK+DR
Sbjct: 533 YGLRPILLLNKVDR 574
>TC86809 homologue to PIR|S35701|S35701 translation elongation factor EF-G
chloroplast - soybean, partial (91%)
Length = 2674
Score = 100 bits (248), Expect = 1e-21
Identities = 56/145 (38%), Positives = 84/145 (57%), Gaps = 4/145 (2%)
Frame = +2
Query: 4 STSDNNDR----KKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDE 59
ST D R K RNI I+AH+D GKTT ++++ +V MD++++
Sbjct: 275 STPDEAKRAVPLKDYRNIGIMAHIDAGKTTTTERILFYTGRNYKIGEVHEGTATMDWMEQ 454
Query: 60 EQRRAITMKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQ 119
EQ R IT+ S++ + +++H +N+ID+PGH+DF EV A R+ DGA+ L D+V GV Q
Sbjct: 455 EQERGITITSAATTTFWDNHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEPQ 634
Query: 120 THAVLRQCWTEMLEPCLVLNKMDRL 144
+ V RQ + +NKMDRL
Sbjct: 635 SETVWRQADRYGVPRICFVNKMDRL 709
>TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein LepA
homolog {Arabidopsis thaliana}, partial (40%)
Length = 1035
Score = 99.8 bits (247), Expect = 2e-21
Identities = 68/178 (38%), Positives = 96/178 (53%), Gaps = 10/178 (5%)
Frame = +1
Query: 2 EESTSDNNDR------KKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMD 55
+ D DR IRN I+AH+DHGK+TLAD+L+ G V P+ K +F+D
Sbjct: 205 DSPAKDGQDRLSKVPVSNIRNFSIIAHIDHGKSTLADKLLQVT--GTV-PQREMKEQFLD 375
Query: 56 YLDEEQRRAITMKSSSISLHY----NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVD 111
+D E+ R IT+K + + Y + +NLID+PGH+DF EVS + +GALL+VD
Sbjct: 376 NMDLERERGITIKLQAARMRYVFENEPYCLNLIDTPGHVDFSYEVSRSLAACEGALLVVD 555
Query: 112 AVEGVHIQTHAVLRQCWTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGI 169
A +GV QT A + LE VLNK+D P R+L+ + EV G+
Sbjct: 556 ASQGVEAQTLANVYLALDNNLEIIPVLNKID--------LPGAEPDRVLKEIEEVIGL 705
>TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (27%)
Length = 672
Score = 97.4 bits (241), Expect = 8e-21
Identities = 56/133 (42%), Positives = 77/133 (57%)
Frame = +3
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
R IRNI I+AHVDHGKTTL D ++ + + R MD D E+ R IT+ S
Sbjct: 177 RADIRNIAIVAHVDHGKTTLVDAMLKQTK--VFRDNQTVQERIMDSNDLERERGITILSK 350
Query: 71 SISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWTE 130
+ S+ Y +N+ID+PGH DF GEV + +G LL+VD+VEG QT VL++
Sbjct: 351 NTSVTYKDAKINIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMPQTRFVLKKALEF 530
Query: 131 MLEPCLVLNKMDR 143
+V+NK+DR
Sbjct: 531 GHAVVVVVNKIDR 569
>TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (98%)
Length = 2589
Score = 97.4 bits (241), Expect = 8e-21
Identities = 56/133 (42%), Positives = 77/133 (57%)
Frame = +3
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
R IRNI I+AHVDHGKTTL D ++ + + R MD D E+ R IT+ S
Sbjct: 387 RADIRNIAIVAHVDHGKTTLVDAMLKQTK--VFRDNQTVQERIMDSNDLERERGITILSK 560
Query: 71 SISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWTE 130
+ S+ Y +N+ID+PGH DF GEV + +G LL+VD+VEG QT VL++
Sbjct: 561 NTSVTYKDAKINIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMPQTRFVLKKALEF 740
Query: 131 MLEPCLVLNKMDR 143
+V+NK+DR
Sbjct: 741 GHAVVVVVNKIDR 779
>TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu chloroplast
precursor (EF-Tu). [Garden pea] {Pisum sativum}, partial
(92%)
Length = 1907
Score = 71.2 bits (173), Expect = 6e-13
Identities = 68/197 (34%), Positives = 97/197 (48%), Gaps = 11/197 (5%)
Frame = +3
Query: 10 DRKKIR-NICILAHVDHGKTTLADQL-IAAASGGMVHPKVAGKVRFMDYLDEEQRRAITM 67
+RKK NI + HVDHGKTTL L +A AS G PK K +D EE+ R IT+
Sbjct: 402 ERKKPHLNIGTIGHVDHGKTTLTAALTMALASLGNSAPK---KYDEIDAAPEERARGITI 572
Query: 68 KSSSISL-----HYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQT-- 120
++++ HY H +D PGH D+ + T A DGA+L+V +G QT
Sbjct: 573 NTATVEYETETRHYAH-----VDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTKE 737
Query: 121 HAVLRQCWTEMLEPCLV--LNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYNSEKY 178
H +L + ++ P +V LNK D++ E L +E R L +E G S
Sbjct: 738 HILLAK---QVGVPSVVVFLNKQDQVDDEELLELVELEVRELLSSYEFPGDDIPIVSGSA 908
Query: 179 LSDVDALLAGGTAAGGE 195
L ++AL+A T G+
Sbjct: 909 LLALEALVANPTIKRGD 959
>TC84431 similar to GP|10176983|dbj|BAB10215. GTP-binding membrane protein
LepA homolog {Arabidopsis thaliana}, partial (46%)
Length = 976
Score = 69.3 bits (168), Expect = 2e-12
Identities = 44/131 (33%), Positives = 69/131 (52%), Gaps = 15/131 (11%)
Frame = +3
Query: 28 TTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSSSISLHY----------- 76
TTLAD+L+ K G+ +++D L E+ R IT+K+ + ++ Y
Sbjct: 3 TTLADRLLELTG---TIKKGLGQPQYLDKLQVERERGITVKAQTATMFYKNIINGDDFKD 173
Query: 77 ----NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWTEML 132
+++ +NLID+PGH+DF EVS + G LL+VDA +GV QT A + L
Sbjct: 174 GKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNL 353
Query: 133 EPCLVLNKMDR 143
V+NK+D+
Sbjct: 354 AIIPVINKIDQ 386
>TC78051 homologue to PIR|T01400|T01400 translation elongation factor EF-Tu
precursor mitochondrial - Arabidopsis thaliana, partial
(87%)
Length = 1681
Score = 58.5 bits (140), Expect = 4e-09
Identities = 48/138 (34%), Positives = 70/138 (49%), Gaps = 11/138 (7%)
Frame = +3
Query: 16 NICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRF--MDYLDEEQRRAITMKSSSIS 73
N+ + HVDHGKTTL AA + + A V F +D EE++R IT+ ++ +
Sbjct: 237 NVGTIGHVDHGKTTLT----AAITRVLADEGKAKAVAFDEIDKAPEEKKRGITIATAHVE 404
Query: 74 L-----HYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQT--HAVLRQ 126
HY H +D PGH D+ + T A DG +L+V A +G QT H +L +
Sbjct: 405 YETAKRHYAH-----VDCPGHADYVKNMITGAAQMDGGILVVSAPDGPMPQTKEHILLAR 569
Query: 127 CWTEMLEPCLV--LNKMD 142
++ P LV LNK+D
Sbjct: 570 ---QVGVPSLVCFLNKVD 614
>TC92627 similar to PIR|T49975|T49975 hypothetical protein F12B17.10 -
Arabidopsis thaliana, partial (25%)
Length = 827
Score = 54.7 bits (130), Expect = 6e-08
Identities = 38/113 (33%), Positives = 58/113 (50%), Gaps = 13/113 (11%)
Frame = +2
Query: 16 NICILAHVDHGKTTLADQLI---AAASGGMVHP-----KVAGKVRF-----MDYLDEEQR 62
N+ I+ HVD GK+TL+ +L+ S +H K+ GK F +D EE+
Sbjct: 224 NLAIVGHVDSGKSTLSGRLLHLLGRISKKEMHKYEKEAKLQGKGSFAYAWALDESSEERE 403
Query: 63 RAITMKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEG 115
R ITM + + V ++DSPGH DF + + A +D A+L++DA G
Sbjct: 404 RGITMTVAVAYFDTKKYHVVVLDSPGHKDFIPNMISGATQADAAVLVIDASLG 562
>TC85179 similar to GP|2662343|dbj|BAA23658.1 EF-1 alpha {Oryza sativa},
partial (96%)
Length = 1789
Score = 46.6 bits (109), Expect = 2e-05
Identities = 34/110 (30%), Positives = 48/110 (42%), Gaps = 13/110 (11%)
Frame = +2
Query: 16 NICILAHVDHGKTTLADQLIAAASGGMVH-----PKVAGKVRF--------MDYLDEEQR 62
NI + HVD GK+T A LI G K A ++ +D L E+
Sbjct: 176 NIVFIGHVDSGKSTTAGHLIYKLGGIKKDVIERFEKEAAEINMCSFKYAWVLDKLKAERE 355
Query: 63 RAITMKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDA 112
R IT+ S + LID+PGH DF + T +D A+L+ D+
Sbjct: 356 RGITIDISLSKFETTKYYCTLIDAPGHRDFIKNMITGTSQADCAVLVYDS 505
>BQ751083 homologue to GP|13925370|gb elongation factor 2 {Neurospora
crassa}, partial (28%)
Length = 721
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/47 (44%), Positives = 30/47 (63%)
Frame = +3
Query: 317 ELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVEGQKSRISRL 363
E + K+ K +L+AVM +LP +DA+L M++ LP PV QK R L
Sbjct: 6 EDKEKEGKQLLKAVMRTFLPAADALLEMMIIHLPSPVTAQKYRSETL 146
>TC89396 similar to SP|P48599|IFE1_ORYSA Eukaryotic translation initiation
factor 4E (EIF-4E) (EIF4E) (mRNA cap-binding protein),
partial (81%)
Length = 915
Score = 32.3 bits (72), Expect = 0.32
Identities = 14/40 (35%), Positives = 21/40 (52%)
Frame = +2
Query: 252 WGPWYYNPKTKMIVGKKGISGSKARPMFVQFVLEPLWQVY 291
W W+ NP+TK K+ GS RP++ +E W +Y
Sbjct: 224 WTFWFDNPQTK---SKQQAWGSSIRPVYTFSTVEEFWSIY 334
>AW691530 similar to PIR|T01917|T01 translation initiation factor IF-2
homolog F2P3.9 - Arabidopsis thaliana, partial (22%)
Length = 581
Score = 30.0 bits (66), Expect = 1.6
Identities = 21/80 (26%), Positives = 34/80 (42%), Gaps = 4/80 (5%)
Frame = +3
Query: 17 ICILAHVDHGKTTLADQL----IAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSSSI 72
+ ++ HVDHGKT+L D L +AA G + + V + M S +
Sbjct: 300 VTVMGHVDHGKTSLLDALRQTSVAAKEAGGITQHLGAFV-------------VGMSSGA- 437
Query: 73 SLHYNHHTVNLIDSPGHIDF 92
++ +D+PGH F
Sbjct: 438 -------SITFLDTPGHAAF 476
>AW695818 similar to PIR|T01917|T01 translation initiation factor IF-2
homolog F2P3.9 - Arabidopsis thaliana, partial (20%)
Length = 594
Score = 30.0 bits (66), Expect = 1.6
Identities = 21/80 (26%), Positives = 34/80 (42%), Gaps = 4/80 (5%)
Frame = +3
Query: 17 ICILAHVDHGKTTLADQL----IAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSSSI 72
+ ++ HVDHGKT+L D L +AA G + + V + M S +
Sbjct: 375 VTVMGHVDHGKTSLLDALRQTSVAAKEAGGITQHLGAFV-------------VGMSSGA- 512
Query: 73 SLHYNHHTVNLIDSPGHIDF 92
++ +D+PGH F
Sbjct: 513 -------SITFLDTPGHAAF 551
>AW685360 homologue to PIR|T00956|T00 translation initiation factor eIF-2
gamma chain F20D22.6 - Arabidopsis thaliana, partial
(37%)
Length = 650
Score = 29.3 bits (64), Expect = 2.7
Identities = 14/34 (41%), Positives = 19/34 (55%)
Frame = +1
Query: 81 VNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVE 114
V+ +D PGH + A + DGALLL+ A E
Sbjct: 475 VSFVDCPGHDILMATMLNGAAIMDGALLLIAANE 576
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,145,175
Number of Sequences: 36976
Number of extensions: 140843
Number of successful extensions: 767
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 739
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 743
length of query: 392
length of database: 9,014,727
effective HSP length: 98
effective length of query: 294
effective length of database: 5,391,079
effective search space: 1584977226
effective search space used: 1584977226
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146552.2


