

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146551.12 + phase: 0 /pseudo
(447 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
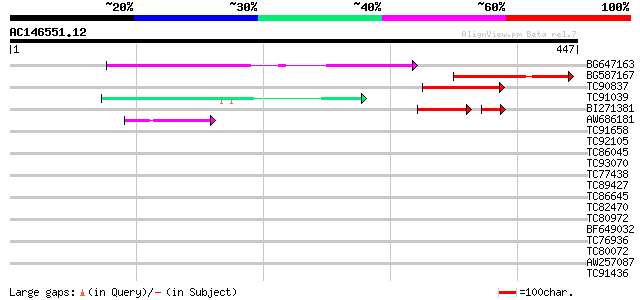
Score E
Sequences producing significant alignments: (bits) Value
BG647163 similar to GP|9757943|dbj emb|CAB72466.1~gene_id:MJC20.... 122 3e-28
BG587167 similar to PIR|T47439|T474 hypothetical protein T18B22.... 98 6e-21
TC90837 weakly similar to GP|22202752|dbj|BAC07409. hypothetical... 64 1e-10
TC91039 similar to PIR|C96640|C96640 hypothetical protein T25B24... 55 7e-08
BI271381 similar to GP|21397269|gb Unknown protein {Oryza sativa... 48 2e-06
AW686181 41 8e-04
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 41 0.001
TC92105 homologue to GP|16197625|gb|AAL13436.1 anaphase promotin... 40 0.001
TC86045 similar to PIR|T46225|T46225 alpha NAC-like protein - Ar... 40 0.002
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 39 0.003
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 39 0.005
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 38 0.007
TC86645 similar to PIR|H86321|H86321 hypothetical protein AAF271... 38 0.009
TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical prot... 37 0.012
TC80972 weakly similar to GP|23496972|gb|AAN36523.1 hypothetical... 36 0.026
BF649032 weakly similar to PIR|T45951|T459 hypothetical protein ... 36 0.026
TC76936 similar to GP|20160782|dbj|BAB89723. putative nascent po... 36 0.034
TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Ar... 36 0.034
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 35 0.045
TC91436 weakly similar to GP|21593499|gb|AAM65466.1 putative nod... 35 0.059
>BG647163 similar to GP|9757943|dbj emb|CAB72466.1~gene_id:MJC20.8~similar to
unknown protein {Arabidopsis thaliana}, partial (51%)
Length = 744
Score = 122 bits (306), Expect = 3e-28
Identities = 82/245 (33%), Positives = 117/245 (47%)
Frame = +3
Query: 77 YIHKSLNGDSEIFRQVYRMYPDVFRKLCMIIREKTPLVDTRFICIEEMLASFLQIVGQNT 136
+ + LNG ++ + +RM VF KLC I+ K L DT I IEE LA FL I+G N
Sbjct: 42 FFEEVLNGPNQRCLENFRMDKVVFYKLCDILETKGLLRDTNRIKIEEQLAIFLFIIGHNL 221
Query: 137 RYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSSVPAKIRESTRFYPYFKILY 196
R ++ F S S +F+ +L A+ SI+ + P V + I E RF+P
Sbjct: 222 RIRGVQELFHYSGETISRHFNNVLNAVMSISKEYFQPPGEDVASMIAEDDRFFP------ 383
Query: 197 N*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLKRACKNKGKHKILSLLQAIDGTHI 256
+ +D G AIDG ++
Sbjct: 384 ---------------YFKDCVG-------------------------------AIDGIYV 425
Query: 257 PALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDALKRKNGL 316
P V + +R++ G +SQNVLAAC+FDL+F YVL+GWEGSA + ++ N A+ RKN L
Sbjct: 426 PVTVGVDEQGPFRNKDGLLSQNVLAACSFDLKFCYVLAGWEGSASNLQVFNSAITRKNKL 605
Query: 317 KVPHG 321
+VP G
Sbjct: 606 QVPEG 620
>BG587167 similar to PIR|T47439|T474 hypothetical protein T18B22.40 -
Arabidopsis thaliana, partial (11%)
Length = 788
Score = 98.2 bits (243), Expect = 6e-21
Identities = 50/94 (53%), Positives = 62/94 (65%)
Frame = +2
Query: 351 FKSRFTIFKSAPPFLFKTQAELVLACAALHNFLRKECRSDEFPIEPSNESSSSSSMLPIH 410
FKS F IFKSAPPF +KT+ E+VLAC LHNFLRK CR+D FP E +E +
Sbjct: 512 FKSLFLIFKSAPPFPYKTKKEIVLACVGLHNFLRKFCRTDNFPEEKDSEDVEDVKEV--- 682
Query: 411 EDNNDELNVQTQEQEREDANIWRTNLGLDMWRDV 444
+NNDE + TQ+Q+RE AN R + DMWRD+
Sbjct: 683 -NNNDE*ILVTQDQQREHANQVRATIATDMWRDL 781
>TC90837 weakly similar to GP|22202752|dbj|BAC07409. hypothetical
protein~similar to Oryza sativa chromosome10
OSJNBa0079H13.17, partial (7%)
Length = 596
Score = 63.9 bits (154), Expect = 1e-10
Identities = 28/65 (43%), Positives = 41/65 (63%)
Frame = +1
Query: 326 PENEKELFNLRHASLRNVVERIFGIFKSRFTIFKSAPPFLFKTQAELVLACAALHNFLRK 385
P N +E FN H+SLR+ +ER FG+ K R+ I P F KTQ ++++A ALHN++R
Sbjct: 106 PTNAQEAFNRAHSSLRSCIERSFGVLKKRWKILNKMPQFSVKTQIDVIIAAFALHNYIRI 285
Query: 386 ECRSD 390
+ D
Sbjct: 286 NSQDD 300
>TC91039 similar to PIR|C96640|C96640 hypothetical protein T25B24.14
[imported] - Arabidopsis thaliana, partial (4%)
Length = 732
Score = 54.7 bits (130), Expect = 7e-08
Identities = 50/219 (22%), Positives = 83/219 (37%), Gaps = 10/219 (4%)
Frame = +1
Query: 73 KGYDYIHKSLNGDSEIFRQVYRMYPDVFRKLCMIIREKTPLVDTRFICIEEMLASFLQIV 132
+G D++++ LNG +RM P +F++LC ++ K L ++ + +EE + F+ +
Sbjct: 229 RGQDWMNEILNGHPVRCMNAFRMDPTLFKQLCEDLQSKYGLQPSKRMTVEEKVGIFVYTL 408
Query: 133 GQNTRYCIIRNTFGRSQFAASENFHKILKALNS-------IAPDLMAK--PTSS-VPAKI 182
+R F S S FH++L+A++ +A D++ PT +P I
Sbjct: 409 AMGASNRDVRERFQHSGETISRAFHEVLEAISGRSRGYRGLARDIIRPKDPTFQFIPLHI 588
Query: 183 RESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLKRACKNKGKH 242
R+ PYFK
Sbjct: 589 SNDERYMPYFK------------------------------------------------- 621
Query: 243 KILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLA 281
+ IDGTHI A + D YR R G + NV+A
Sbjct: 622 ---DCIGCIDGTHIAACIPEADQMRYRGRKGIPTFNVMA 729
>BI271381 similar to GP|21397269|gb Unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 407
Score = 47.8 bits (112), Expect(2) = 2e-06
Identities = 20/43 (46%), Positives = 28/43 (64%)
Frame = +2
Query: 322 HGNDPENEKELFNLRHASLRNVVERIFGIFKSRFTIFKSAPPF 364
H ++ +ELFN RH+SLR +ER FG+ K+RF I K P +
Sbjct: 113 HNQQDKSPEELFNYRHSSLRMTIERCFGVLKNRFPILKLMPSY 241
Score = 21.9 bits (45), Expect(2) = 2e-06
Identities = 9/19 (47%), Positives = 12/19 (62%)
Frame = +3
Query: 373 VLACAALHNFLRKECRSDE 391
V AC A+HN++ K DE
Sbjct: 264 VTACCAIHNYICKWNLPDE 320
>AW686181
Length = 304
Score = 41.2 bits (95), Expect = 8e-04
Identities = 23/72 (31%), Positives = 40/72 (54%)
Frame = +3
Query: 91 QVYRMYPDVFRKLCMIIREKTPLVDTRFICIEEMLASFLQIVGQNTRYCIIRNTFGRSQF 150
+++RM +F KLC + E L ++ + +EEM+A FL +VG +I+ F S
Sbjct: 3 EMFRMEKHIFHKLCHELVEHD-LKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGE 179
Query: 151 AASENFHKILKA 162
S +F+++L A
Sbjct: 180 TVSRHFYRVLHA 215
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 40.8 bits (94), Expect = 0.001
Identities = 16/28 (57%), Positives = 22/28 (78%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
+D D + DDDDDD+DDDD +DE+ +EE
Sbjct: 92 DDEDGDEEDDDDDDDDDDDEEDEDDEEE 175
Score = 40.0 bits (92), Expect = 0.002
Identities = 15/28 (53%), Positives = 22/28 (78%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
+DI+ + D+D D+EDDDD DD++ DEE
Sbjct: 71 DDIEVDEDDEDGDEEDDDDDDDDDDDEE 154
Score = 38.9 bits (89), Expect = 0.004
Identities = 19/28 (67%), Positives = 21/28 (74%), Gaps = 1/28 (3%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDD-DDTDDEETDE 29
ED D E DDDDDD+DD +D DDEE DE
Sbjct: 98 EDGDEEDDDDDDDDDDDEEDEDDEEEDE 181
Score = 38.1 bits (87), Expect = 0.007
Identities = 15/27 (55%), Positives = 21/27 (77%)
Frame = +2
Query: 4 DIDTEAIDDDDDDEDDDDTDDEETDEE 30
D D E D++DDD+DDDD DDEE +++
Sbjct: 86 DEDDEDGDEEDDDDDDDDDDDEEDEDD 166
Score = 37.7 bits (86), Expect = 0.009
Identities = 15/30 (50%), Positives = 22/30 (73%)
Frame = +2
Query: 1 MSEDIDTEAIDDDDDDEDDDDTDDEETDEE 30
+ ED + +DDDDD+DDDD +++E DEE
Sbjct: 83 VDEDDEDGDEEDDDDDDDDDDDEEDEDDEE 172
Score = 34.3 bits (77), Expect = 0.10
Identities = 11/27 (40%), Positives = 21/27 (77%)
Frame = +2
Query: 4 DIDTEAIDDDDDDEDDDDTDDEETDEE 30
++D + D D++D+DDDD DD++ ++E
Sbjct: 80 EVDEDDEDGDEEDDDDDDDDDDDEEDE 160
>TC92105 homologue to GP|16197625|gb|AAL13436.1 anaphase promoting complex
subunit 11 {Arabidopsis thaliana}, partial (98%)
Length = 549
Score = 40.4 bits (93), Expect = 0.001
Identities = 15/21 (71%), Positives = 17/21 (80%)
Frame = +3
Query: 282 ACNFDLEFMYVLSGWEGSAHD 302
AC+FD EFMY LSGW+ S HD
Sbjct: 462 ACSFDFEFMYALSGWKSSTHD 524
>TC86045 similar to PIR|T46225|T46225 alpha NAC-like protein - Arabidopsis
thaliana, partial (69%)
Length = 1166
Score = 39.7 bits (91), Expect = 0.002
Identities = 15/29 (51%), Positives = 22/29 (75%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEEL 31
ED+ + DD+DDDEDDDD DD++ ++ L
Sbjct: 220 EDVKDDDKDDEDDDEDDDDEDDDDKEDAL 306
Score = 37.4 bits (85), Expect = 0.012
Identities = 14/25 (56%), Positives = 20/25 (80%)
Frame = +1
Query: 11 DDDDDDEDDDDTDDEETDEELYEAI 35
DDD DDEDDD+ DD+E D++ +A+
Sbjct: 232 DDDKDDEDDDEDDDDEDDDDKEDAL 306
Score = 32.7 bits (73), Expect = 0.29
Identities = 13/28 (46%), Positives = 19/28 (67%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
E+ D I+D DD+ DD+ DDE+ D+E
Sbjct: 196 EEDDAPIIEDVKDDDKDDEDDDEDDDDE 279
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 39.3 bits (90), Expect = 0.003
Identities = 15/28 (53%), Positives = 22/28 (78%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
ED + E D+D+DD+DDDD DDE+ D++
Sbjct: 253 EDDEDEDEDEDEDDDDDDDDDDEDDDDD 336
Score = 38.5 bits (88), Expect = 0.005
Identities = 14/28 (50%), Positives = 23/28 (82%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
E++D E +D+D+DED+DD DD++ D+E
Sbjct: 238 EEMDFEDDEDEDEDEDEDDDDDDDDDDE 321
Score = 35.8 bits (81), Expect = 0.034
Identities = 13/30 (43%), Positives = 22/30 (73%)
Frame = +1
Query: 1 MSEDIDTEAIDDDDDDEDDDDTDDEETDEE 30
M + D + +D+D+D+DDDD DD+E D++
Sbjct: 244 MDFEDDEDEDEDEDEDDDDDDDDDDEDDDD 333
Score = 35.8 bits (81), Expect = 0.034
Identities = 13/31 (41%), Positives = 22/31 (70%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEELYE 33
+D D + +D+DDD+DDDD D+++ D+ E
Sbjct: 256 DDEDEDEDEDEDDDDDDDDDDEDDDDDGFAE 348
Score = 33.9 bits (76), Expect = 0.13
Identities = 15/26 (57%), Positives = 18/26 (68%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETD 28
ED D + DDDDD++DDDD E TD
Sbjct: 280 EDEDDDDDDDDDDEDDDDDGFAEPTD 357
Score = 32.0 bits (71), Expect = 0.50
Identities = 9/28 (32%), Positives = 22/28 (78%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
++I+ +DD+D+++D+D DD++ D++
Sbjct: 229 DEIEEMDFEDDEDEDEDEDEDDDDDDDD 312
Score = 30.0 bits (66), Expect = 1.9
Identities = 11/28 (39%), Positives = 22/28 (78%), Gaps = 1/28 (3%)
Frame = +1
Query: 4 DIDTEAID-DDDDDEDDDDTDDEETDEE 30
D + E +D +DD+DED+D+ +D++ D++
Sbjct: 226 DDEIEEMDFEDDEDEDEDEDEDDDDDDD 309
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 - garden
pea, partial (93%)
Length = 1607
Score = 38.5 bits (88), Expect = 0.005
Identities = 14/28 (50%), Positives = 23/28 (82%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
+D D +A DDD+D+++D+D DD+E +EE
Sbjct: 1049 DDEDDDAEDDDEDEDEDEDEDDDEDEEE 1132
Score = 38.5 bits (88), Expect = 0.005
Identities = 15/27 (55%), Positives = 21/27 (77%)
Frame = +2
Query: 4 DIDTEAIDDDDDDEDDDDTDDEETDEE 30
D+D E D+DDD EDDD+ +DE+ DE+
Sbjct: 1031 DLDDEDDDEDDDAEDDDEDEDEDEDED 1111
Score = 35.4 bits (80), Expect = 0.045
Identities = 12/28 (42%), Positives = 22/28 (77%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
+D + + +DDD+DED+D+ +D++ DEE
Sbjct: 1046 DDDEDDDAEDDDEDEDEDEDEDDDEDEE 1129
Score = 35.0 bits (79), Expect = 0.059
Identities = 13/30 (43%), Positives = 22/30 (73%)
Frame = +2
Query: 1 MSEDIDTEAIDDDDDDEDDDDTDDEETDEE 30
+ ++ D E D +DDDED+D+ +DE+ DE+
Sbjct: 1034 LDDEDDDEDDDAEDDDEDEDEDEDEDDDED 1123
Score = 32.7 bits (73), Expect = 0.29
Identities = 11/25 (44%), Positives = 20/25 (80%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEET 27
+D + + D+D+D+++DDD D+EET
Sbjct: 1061 DDAEDDDEDEDEDEDEDDDEDEEET 1135
Score = 30.4 bits (67), Expect = 1.4
Identities = 10/28 (35%), Positives = 21/28 (74%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
ED D + +DDD+++D+D D+++ ++E
Sbjct: 1043 EDDDEDDDAEDDDEDEDEDEDEDDDEDE 1126
Score = 28.9 bits (63), Expect = 4.2
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +2
Query: 8 EAIDDDDDDEDDDDTDDEETDEE 30
E D DD++DD+D D E+ DE+
Sbjct: 1019 EEFGDLDDEDDDEDDDAEDDDED 1087
Score = 27.7 bits (60), Expect = 9.4
Identities = 9/25 (36%), Positives = 19/25 (76%)
Frame = +2
Query: 6 DTEAIDDDDDDEDDDDTDDEETDEE 30
D E D+D+D++DD+D ++ +T ++
Sbjct: 1076 DDEDEDEDEDEDDDEDEEETKTKKK 1150
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 38.1 bits (87), Expect = 0.007
Identities = 15/25 (60%), Positives = 20/25 (80%)
Frame = +1
Query: 6 DTEAIDDDDDDEDDDDTDDEETDEE 30
D + DDDDDD+D+DD +DE+ DEE
Sbjct: 448 DDDDEDDDDDDDDNDDGEDEDEDEE 522
Score = 35.8 bits (81), Expect = 0.034
Identities = 14/28 (50%), Positives = 21/28 (75%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
+D D + DDD+DD +D+D D+EE D+E
Sbjct: 454 DDEDDDDDDDDNDDGEDEDEDEEEDDDE 537
Score = 35.4 bits (80), Expect = 0.045
Identities = 13/28 (46%), Positives = 20/28 (71%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEE 30
E+ D +DDDD++DDDD +DE+ D +
Sbjct: 286 ENKDGSETEDDDDEDDDDDVNDEDDDND 369
Score = 35.0 bits (79), Expect = 0.059
Identities = 14/27 (51%), Positives = 19/27 (69%)
Frame = +1
Query: 4 DIDTEAIDDDDDDEDDDDTDDEETDEE 30
D D E DDDDDD DD + +DE+ +E+
Sbjct: 448 DDDDEDDDDDDDDNDDGEDEDEDEEED 528
Score = 33.5 bits (75), Expect = 0.17
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 7/36 (19%)
Frame = +1
Query: 2 SEDIDTEAIDDDDDDEDDD-------DTDDEETDEE 30
+ED D E DDD +DEDDD D DDE+ D E
Sbjct: 307 TEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPE 414
Score = 32.0 bits (71), Expect = 0.50
Identities = 11/24 (45%), Positives = 19/24 (78%)
Frame = +1
Query: 7 TEAIDDDDDDEDDDDTDDEETDEE 30
+E DDDD+D+DDD D+++ ++E
Sbjct: 301 SETEDDDDEDDDDDVNDEDDDNDE 372
Score = 31.6 bits (70), Expect = 0.65
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 5/33 (15%)
Frame = +1
Query: 2 SEDIDTEAIDDDDD-----DEDDDDTDDEETDE 29
S+D D + DDDDD DED+D+ +D++ D+
Sbjct: 445 SDDDDEDDDDDDDDNDDGEDEDEDEEEDDDEDQ 543
Score = 30.4 bits (67), Expect = 1.4
Identities = 13/25 (52%), Positives = 17/25 (68%)
Frame = +1
Query: 2 SEDIDTEAIDDDDDDEDDDDTDDEE 26
SE D + DDDDD D+DD +DE+
Sbjct: 301 SETEDDDDEDDDDDVNDEDDDNDED 375
Score = 30.4 bits (67), Expect = 1.4
Identities = 11/24 (45%), Positives = 18/24 (74%)
Frame = +1
Query: 6 DTEAIDDDDDDEDDDDTDDEETDE 29
+TE DD+DDD+D +D DD+ ++
Sbjct: 304 ETEDDDDEDDDDDVNDEDDDNDED 375
Score = 27.7 bits (60), Expect = 9.4
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 7/36 (19%)
Frame = +1
Query: 2 SEDIDTEAIDDDDDD-------EDDDDTDDEETDEE 30
S D D E D +DD DDDD DD++ D++
Sbjct: 379 SGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDD 486
>TC86645 similar to PIR|H86321|H86321 hypothetical protein AAF27100.1
[imported] - Arabidopsis thaliana, partial (76%)
Length = 1116
Score = 37.7 bits (86), Expect = 0.009
Identities = 18/32 (56%), Positives = 23/32 (71%), Gaps = 3/32 (9%)
Frame = +3
Query: 2 SEDIDTEAID---DDDDDEDDDDTDDEETDEE 30
+E+ D EA D DDDD EDDDD +D++ DEE
Sbjct: 738 AEEDDDEAGDAGKDDDDSEDDDDQEDDDDDEE 833
Score = 32.0 bits (71), Expect = 0.50
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = +3
Query: 6 DTEAIDDDDDDEDDDDTDDEE 26
D + +DDDD EDDDD ++EE
Sbjct: 777 DDDDSEDDDDQEDDDDDEEEE 839
Score = 30.8 bits (68), Expect = 1.1
Identities = 10/20 (50%), Positives = 16/20 (80%)
Frame = +3
Query: 11 DDDDDDEDDDDTDDEETDEE 30
DDD +D+DD + DD++ +EE
Sbjct: 780 DDDSEDDDDQEDDDDDEEEE 839
>TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (8%)
Length = 1064
Score = 37.4 bits (85), Expect = 0.012
Identities = 15/24 (62%), Positives = 18/24 (74%)
Frame = +2
Query: 6 DTEAIDDDDDDEDDDDTDDEETDE 29
D + DDDDDD+DDDD DD TD+
Sbjct: 35 DDDDDDDDDDDDDDDDDDDMSTDK 106
Score = 36.2 bits (82), Expect = 0.026
Identities = 12/20 (60%), Positives = 18/20 (90%)
Frame = +2
Query: 12 DDDDDEDDDDTDDEETDEEL 31
DDDDD+DDDD DD++ D+++
Sbjct: 35 DDDDDDDDDDDDDDDDDDDM 94
Score = 35.0 bits (79), Expect = 0.059
Identities = 12/25 (48%), Positives = 19/25 (76%)
Frame = +2
Query: 11 DDDDDDEDDDDTDDEETDEELYEAI 35
DDDDDD+DDDD DD+ + ++ + +
Sbjct: 47 DDDDDDDDDDDDDDDMSTDKSFSTL 121
Score = 33.9 bits (76), Expect = 0.13
Identities = 14/35 (40%), Positives = 22/35 (62%)
Frame = +2
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEELYEAIYM 37
+D D + DDDDDD+DDDD +++ L E + +
Sbjct: 35 DDDDDDDDDDDDDDDDDDDMSTDKSFSTLKEKLVL 139
>TC80972 weakly similar to GP|23496972|gb|AAN36523.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (9%)
Length = 662
Score = 36.2 bits (82), Expect = 0.026
Identities = 17/38 (44%), Positives = 27/38 (70%), Gaps = 1/38 (2%)
Frame = -2
Query: 1 MSEDIDTEAIDDDDD-DEDDDDTDDEETDEELYEAIYM 37
+ ED D +A DD+++ DE++DD +EE EELY +Y+
Sbjct: 631 IEEDDDDDADDDNEEKDEEEDDEVEEEEQEELYG*VYV 518
>BF649032 weakly similar to PIR|T45951|T459 hypothetical protein F7J8.90 -
Arabidopsis thaliana, partial (11%)
Length = 387
Score = 36.2 bits (82), Expect = 0.026
Identities = 19/39 (48%), Positives = 24/39 (60%), Gaps = 3/39 (7%)
Frame = -3
Query: 386 ECRSDE---FPIEPSNESSSSSSMLPIHEDNNDELNVQT 421
EC++ E FP P N SS+SSS+LP+H N L V T
Sbjct: 370 ECKNQEKXPFPPSPXNSSSASSSILPLHNPFNRVLYVTT 254
>TC76936 similar to GP|20160782|dbj|BAB89723. putative nascent polypeptide
associated complex alpha chain {Oryza sativa (japonica
cultivar-group)}, partial (86%)
Length = 914
Score = 35.8 bits (81), Expect = 0.034
Identities = 11/25 (44%), Positives = 20/25 (80%)
Frame = +3
Query: 6 DTEAIDDDDDDEDDDDTDDEETDEE 30
D ++DDDD++D+DD DD++ D++
Sbjct: 138 DEPVVEDDDDEDDEDDEDDDDEDDD 212
Score = 33.1 bits (74), Expect = 0.22
Identities = 15/26 (57%), Positives = 18/26 (68%)
Frame = +3
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETD 28
ED D E DD+DDEDDDD DD+ +
Sbjct: 153 EDDDDE---DDEDDEDDDDEDDDNAE 221
Score = 31.2 bits (69), Expect = 0.85
Identities = 10/23 (43%), Positives = 19/23 (82%)
Frame = +3
Query: 11 DDDDDDEDDDDTDDEETDEELYE 33
DD+DD++D+DD D+++ + E +E
Sbjct: 162 DDEDDEDDEDDDDEDDDNAEGFE 230
Score = 30.4 bits (67), Expect = 1.4
Identities = 12/26 (46%), Positives = 18/26 (69%)
Frame = +3
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETD 28
+D + + D+DDDDEDDD+ + E D
Sbjct: 159 DDDEDDEDDEDDDDEDDDNAEGFEGD 236
Score = 28.5 bits (62), Expect = 5.5
Identities = 10/21 (47%), Positives = 16/21 (75%)
Frame = +3
Query: 1 MSEDIDTEAIDDDDDDEDDDD 21
+ +D D + DD+DDD++DDD
Sbjct: 150 VEDDDDEDDEDDEDDDDEDDD 212
>TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 746
Score = 35.8 bits (81), Expect = 0.034
Identities = 13/26 (50%), Positives = 20/26 (76%)
Frame = +1
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETD 28
+DID + D+D D+E+DDD +D+E D
Sbjct: 472 KDIDNDETDEDGDEEEDDDDEDDEYD 549
Score = 29.3 bits (64), Expect = 3.2
Identities = 9/24 (37%), Positives = 19/24 (78%)
Frame = +1
Query: 10 IDDDDDDEDDDDTDDEETDEELYE 33
ID+D+ DED D+ +D++ +++ Y+
Sbjct: 478 IDNDETDEDGDEEEDDDDEDDEYD 549
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 35.4 bits (80), Expect = 0.045
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 3/35 (8%)
Frame = -3
Query: 2 SEDIDTEAIDDDDDDED---DDDTDDEETDEELYE 33
S + +DD+D+DED D+D D++E DEEL E
Sbjct: 407 SSSSSSSLLDDEDEDEDEDEDEDEDEDEDDEELLE 303
Score = 32.3 bits (72), Expect = 0.38
Identities = 13/27 (48%), Positives = 20/27 (73%)
Frame = -3
Query: 4 DIDTEAIDDDDDDEDDDDTDDEETDEE 30
D + E D+D+D+++D+D DDEE EE
Sbjct: 380 DDEDEDEDEDEDEDEDEDEDDEELLEE 300
Score = 31.2 bits (69), Expect = 0.85
Identities = 12/32 (37%), Positives = 22/32 (68%)
Frame = -3
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEELYEA 34
+D D + +D+D+DED+D+ D+E +E E+
Sbjct: 380 DDEDEDEDEDEDEDEDEDEDDEELLEESESES 285
Score = 29.6 bits (65), Expect = 2.5
Identities = 13/42 (30%), Positives = 25/42 (58%)
Frame = -3
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEELYEAIYMYMMAIHA 44
ED D + +D+D+DEDD++ +E E ++ ++ +A A
Sbjct: 368 EDEDEDEDEDEDEDEDDEELLEESESESESDSSFLAGVAFAA 243
Score = 29.6 bits (65), Expect = 2.5
Identities = 13/40 (32%), Positives = 23/40 (57%)
Frame = -3
Query: 3 EDIDTEAIDDDDDDEDDDDTDDEETDEELYEAIYMYMMAI 42
ED D + +D+D+DED+DD + E E E+ ++ +
Sbjct: 374 EDEDEDEDEDEDEDEDEDDEELLEESESESESDSSFLAGV 255
Score = 29.3 bits (64), Expect = 3.2
Identities = 9/29 (31%), Positives = 21/29 (72%)
Frame = -3
Query: 2 SEDIDTEAIDDDDDDEDDDDTDDEETDEE 30
S + ++ DD+D+++D+D D++E ++E
Sbjct: 410 SSSSSSSSLLDDEDEDEDEDEDEDEDEDE 324
>TC91436 weakly similar to GP|21593499|gb|AAM65466.1 putative nodulin
protein N21 {Arabidopsis thaliana}, partial (16%)
Length = 909
Score = 35.0 bits (79), Expect = 0.059
Identities = 12/25 (48%), Positives = 19/25 (76%)
Frame = +3
Query: 2 SEDIDTEAIDDDDDDEDDDDTDDEE 26
S+++D ++D DD+DDDD DD+E
Sbjct: 501 SKEVDNNKVEDATDDDDDDDDDDDE 575
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,256,832
Number of Sequences: 36976
Number of extensions: 191814
Number of successful extensions: 4293
Number of sequences better than 10.0: 156
Number of HSP's better than 10.0 without gapping: 1859
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3048
length of query: 447
length of database: 9,014,727
effective HSP length: 99
effective length of query: 348
effective length of database: 5,354,103
effective search space: 1863227844
effective search space used: 1863227844
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146551.12


