

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146549.11 + phase: 0
(597 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
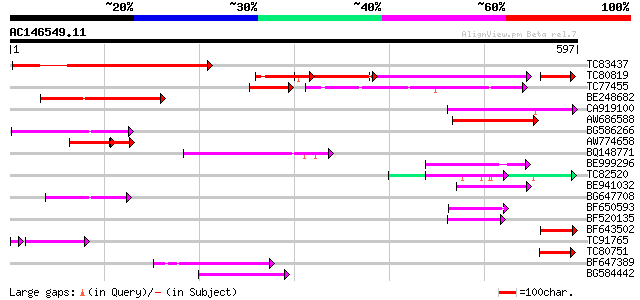
Score E
Sequences producing significant alignments: (bits) Value
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 241 8e-64
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 153 4e-60
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 109 5e-33
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 134 1e-31
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 132 4e-31
AW686588 107 1e-23
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 97 1e-20
AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsut... 58 4e-15
BQ148771 74 1e-13
BE999296 74 1e-13
TC82520 74 2e-13
BE941032 66 4e-11
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 65 1e-10
BF650593 64 2e-10
BF520135 61 1e-09
BF643502 51 1e-06
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 49 3e-06
TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vign... 50 3e-06
BF647389 49 5e-06
BG584442 47 3e-05
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 241 bits (614), Expect = 8e-64
Identities = 125/210 (59%), Positives = 153/210 (72%)
Frame = +2
Query: 4 MGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLM 63
M F LWRKW+KECVSTAT SVLVNGSPT NVLM
Sbjct: 380 MSFLVLWRKWIKECVSTATTSVLVNGSPT---------------------------NVLM 478
Query: 64 TSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKV 123
S V + LFT YS G+ +P V+SHLQFA+DTLLL K+WAN+RALR+ LVIF MSGLKV
Sbjct: 479 KSLVQTQLFTRYSFGVVNPVVVSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKV 658
Query: 124 NYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRSR 183
N++KS LV +NIA SWL EAAS+LS K+GK PF+YLG+PI G++RRL FW+P++ R+++R
Sbjct: 659 NFHKSGLVCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNRIKAR 838
Query: 184 LSEWKSRNLSYGGRLILLKSVLSSLPVYAL 213
L+ W SR LS+GGRL+LLKSVL+SL VYAL
Sbjct: 839 LTGWNSRFLSFGGRLVLLKSVLTSLSVYAL 928
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 153 bits (386), Expect(3) = 4e-60
Identities = 72/170 (42%), Positives = 99/170 (57%)
Frame = +2
Query: 380 EMNILGWGENGEAWRWRRRLLAWEEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTV 439
+M LGW +G AW W RRL WEEE V E ILL N LQD +D W W + +GY+V
Sbjct: 383 DMGRLGWTVDGRAWEWTRRLFVWEEECVRECCILLNNFVLQDNVNDKWRWLLDPVNGYSV 562
Query: 440 RGVYQMLMRQERHNYDVVSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQ 499
+ Y+ + + + D WHK P KVS++ WRLLRNR PTKDNLV RGV+
Sbjct: 563 KVFYRYITSTGHISDRSLVDDVWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVLLATNA 742
Query: 500 FCVTGCGQNETIDLLIIHCPIFGALWQQIKTWIGVFFVDPHQVLDHYYQF 549
CV GC +E+ L +HC +F +LW ++ W+G+ + ++ H+ QF
Sbjct: 743 ACVCGCVDSESTTHLFLHCNVFCSLWSLVRNWLGIPSMSSGELRTHFIQF 892
Score = 81.6 bits (200), Expect(3) = 4e-60
Identities = 38/87 (43%), Positives = 53/87 (60%)
Frame = +3
Query: 301 GGRLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESF 360
GG L EGG S WW+ I K++ GVG G+WFEE+I +G+G + FFW D W G
Sbjct: 144 GGWLCEGGRQSSMWWKTICKVREGVGEGVGNWFEENIRMVVGDGRDAFFWYDTWAGDVPL 323
Query: 361 MVRFRRLYDLSIHKDLSVGEMNILGWG 387
+++ RL DL++ K+ VG + GWG
Sbjct: 324 RLKYPRLLDLAMDKECKVG-LTWGGWG 401
Score = 68.6 bits (166), Expect = 7e-12
Identities = 33/65 (50%), Positives = 44/65 (66%), Gaps = 3/65 (4%)
Frame = +1
Query: 260 GLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGG---RLKEGGMTGSAWWR 316
GLGV FN++LLGKWCWR LVD+EGLW RVL +RYGE+GG R+ + + G +
Sbjct: 31 GLGVGA---FNLSLLGKWCWRLLVDKEGLWHRVLKARYGEEGGGFVRVVDSLLCGGRLYA 201
Query: 317 EIIKI 321
+K+
Sbjct: 202 RCVKV 216
Score = 36.6 bits (83), Expect(3) = 4e-60
Identities = 14/36 (38%), Positives = 23/36 (63%)
Frame = +1
Query: 560 LLEKVKITSLKWLKAKNVCFPFGYHLWRQRPLACLG 595
L++++K+ S WLK+K V F + Y W + L C+G
Sbjct: 1009 LIDRIKLHSFLWLKSKQVGFAYSYLDWWKNSLLCMG 1116
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 109 bits (273), Expect(2) = 5e-33
Identities = 69/245 (28%), Positives = 127/245 (51%), Gaps = 11/245 (4%)
Frame = -2
Query: 312 SAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESFMVRFRRLYDLS 371
S WW++++ ++ V WF + +++G+G ++FFW D W + F R +
Sbjct: 778 SRWWKDLMSLEEVGRVR---WFPRELIRKVGDGRSSFFWKDAWDSSVPLRESFPRAF--F 614
Query: 372 IHKDLSVGEMNILGWGENGEAWR--WRR-RLLAWEEELVVEIRILLTNVTLQDTESDVWL 428
++ L +G ++ G WR WRR L WE+E ++E+ L V L+ +D+W+
Sbjct: 613 PYRLLKMGCGDLWDMNAEGVRWRLYWRRLELFEWEKERLLELLGRLEGVVLR-YWADIWV 437
Query: 429 WRPNIGDGYTVRGVYQML----MRQERHNY--DVVSDAPWHKSAPLKVSIWAWRLLRNRW 482
W+P+ ++V Y +L + ++R +Y +V+ W AP KV ++W L +R
Sbjct: 436 WKPDKEGVFSVNSCYFLLQNLRLLEDRLSYEEEVIFRELWKSKAPAKVLAFSWTLFLDRI 257
Query: 483 PTKDNLVRRGVISY-DTQFCV-TGCGQNETIDLLIIHCPIFGALWQQIKTWIGVFFVDPH 540
PT NL +R ++ D++ CV GC Q+ET+ L +HC + + +++ W+ + P
Sbjct: 256 PTMVNLGKRRLLRVEDSKRCVFCGC-QDETVVHLFLHCDVISKV*REVMRWLNFNLISPP 80
Query: 541 QVLDH 545
+L H
Sbjct: 79 NLLIH 65
Score = 50.1 bits (118), Expect(2) = 5e-33
Identities = 23/46 (50%), Positives = 28/46 (60%)
Frame = -3
Query: 253 CMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYG 298
C+ + GGLGVR + NV+LL KW WR L D+ LW VL YG
Sbjct: 969 CLPRCKGGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYG 832
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 134 bits (337), Expect = 1e-31
Identities = 67/132 (50%), Positives = 95/132 (71%)
Frame = +3
Query: 33 DEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFAD 92
+E +QRGL+QGDPL+PFLFLL AEG++ LM +AVN NLF G+ + T +SHLQ+AD
Sbjct: 6 EEISVQRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDV-KRGGTRVSHLQYAD 182
Query: 93 DTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVGINIAESWLQEAASILSCKLG 152
DTL +G+ + N+ L+++L FE SGLKVN++KS L+GIN+ +++ A L+C+
Sbjct: 183 DTLCIGMPTVDNLWTLKALLQGFEMASGLKVNFHKSSLIGINVPRDFMEAACRFLNCREE 362
Query: 153 KTPFMYLGLPIG 164
PF+YLGLP G
Sbjct: 363 SIPFIYLGLPGG 398
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 132 bits (332), Expect = 4e-31
Identities = 69/169 (40%), Positives = 88/169 (51%), Gaps = 33/169 (19%)
Frame = -2
Query: 462 WHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIF 521
WH+ PLKVS++AWRLLR+R PTK NL+ RGVI + CV+GCG E+ L + C F
Sbjct: 581 WHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGLCVSGCGALESAQHLFLSCSYF 402
Query: 522 GALWQQIKTWIGVFFVDPHQVLDHYYQFVYS----------------------------- 552
+LW ++ WIG VD + + DH+ QFV+S
Sbjct: 401 ASLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGNKASQSFLQLIWLLCAWVLWTERNNM 222
Query: 553 ----SANTTEQLLEKVKITSLKWLKAKNVCFPFGYHLWRQRPLACLGIG 597
S +LL+KVK SL WLKA+N F FG W PL CLGIG
Sbjct: 221 CFNDSITPLPRLLDKVKYLSLGWLKARNASFLFGTFSWWSNPLQCLGIG 75
>AW686588
Length = 567
Score = 107 bits (267), Expect = 1e-23
Identities = 48/90 (53%), Positives = 62/90 (68%)
Frame = +1
Query: 467 PLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQ 526
PLKVSI AWRL+R+R PTK NLVRR ++ + CV GCG ET + L +HC FGA+WQ
Sbjct: 145 PLKVSILAWRLIRDRLPTKANLVRRRCLAVEAAGCVVGCGIAETANHLFLHCATFGAVWQ 324
Query: 527 QIKTWIGVFFVDPHQVLDHYYQFVYSSANT 556
I+ WIGV DPH + DH+ QF+ + +T
Sbjct: 325 HIRAWIGVSGADPHDLSDHFIQFITCTGHT 414
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 97.4 bits (241), Expect = 1e-20
Identities = 50/128 (39%), Positives = 72/128 (56%)
Frame = -3
Query: 3 KMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVL 62
++GF +W W+ ECVST + S L+NG P RGLRQGDPLSP+LF+L E L+ L
Sbjct: 496 RLGFHGIWISWIMECVSTVSYSFLINGGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGL 317
Query: 63 MTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLK 122
A+ G + + P + +HL FADDT+ G + ++ L SI+ + SG
Sbjct: 316 CQQALRKGTLPGVKVARNCPPI-NHLLFADDTMFFGKSNASSCAILLSIMDKYRAASGRC 140
Query: 123 VNYNKSLL 130
+N KS +
Sbjct: 139 IN*TKSAI 116
>AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsutum}, partial
(4%)
Length = 665
Score = 58.2 bits (139), Expect(2) = 4e-15
Identities = 28/48 (58%), Positives = 36/48 (74%)
Frame = -1
Query: 64 TSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSI 111
++ ++ ++ SIGMHS TV SHLQFADDTLLLGVKSWAN +S+
Sbjct: 449 SNLLSHSICLNLSIGMHSLTVFSHLQFADDTLLLGVKSWANAPCGQSL 306
Score = 41.2 bits (95), Expect(2) = 4e-15
Identities = 19/26 (73%), Positives = 23/26 (88%)
Frame = -3
Query: 106 RALRSILVIFENMSGLKVNYNKSLLV 131
RALRSILVIFENMSGLKVN + +++
Sbjct: 327 RALRSILVIFENMSGLKVNLREEVII 250
>BQ148771
Length = 680
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/165 (29%), Positives = 80/165 (47%), Gaps = 7/165 (4%)
Frame = -3
Query: 184 LSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHRK 243
L+ WK+ +LS R+ L KSV+ ++P+Y + P I I+ + KF WG +E R+
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 244 IAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGGR 303
V W T+ K GLG+RRL N A + K W L V+ +Y ++
Sbjct: 393 YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKY-QRSES 217
Query: 304 LKEGGM---TGSAWWREIIK----IQNGVGVEGGSWFEESISKRL 341
L+E + T S+ W+ ++K I+ + G+W E + + L
Sbjct: 216 LEEIFLEKPTDSSLWKALVKLWPEIERNLVDSNGNWNWEKLKQWL 82
>BE999296
Length = 384
Score = 74.3 bits (181), Expect = 1e-13
Identities = 42/110 (38%), Positives = 54/110 (48%)
Frame = -1
Query: 439 VRGVYQMLMRQERHNYDVVSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDT 498
+ G YQ LM + D+ WH PLKV + R+LRN PTKDN VRR VI +
Sbjct: 306 IHGAYQFLMSADAPLDREYIDSVWHNHIPLKVCFFVLRVLRNCLPTKDNFVRRRVIHEEH 127
Query: 499 QFCVTGCGQNETIDLLIIHCPIFGALWQQIKTWIGVFFVDPHQVLDHYYQ 548
C TGC ET D L LW + W+ + V+ + DH+YQ
Sbjct: 126 MLCPTGCSFKETTDDLF--------LWPLV*QWLHIS*VNSCVLRDHFYQ 1
>TC82520
Length = 833
Score = 73.9 bits (180), Expect = 2e-13
Identities = 63/256 (24%), Positives = 90/256 (34%), Gaps = 58/256 (22%)
Frame = +2
Query: 399 LLAWEEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVS 458
L AWEEE V E LL N LQ+ DV W + GYT
Sbjct: 2 LFAWEEESVREWYALLHNTVLQENVHDVCRWLLDPI*GYT-------------------- 121
Query: 459 DAPWHKSAPLKVSIWAW---------RLLRNRWPTKDNLVRRGVIS--------YDTQFC 501
+ +S + W R L + K VR +S ++ C
Sbjct: 122 ------EGNISLSHYLWTTIG*ESS*RCLAKEYSFKGVYVRVASLSQ*TSHEG*FNAATC 283
Query: 502 VT--------GCGQNETIDLLIIHCPIFGALWQQIKTWIGVFFVDPHQVLDHYYQFV--- 550
+ GCG +ET L +HC IFG+LW + W+ + V P + + QF
Sbjct: 284 SSTNSHGLHFGCGDSETATHLFLHCDIFGSLWSHVLRWLHLLLVLPADIRQFFIQFTSMA 463
Query: 551 ------------------------------YSSANTTEQLLEKVKITSLKWLKAKNVCFP 580
+S + +E+VK+ S WLK + F
Sbjct: 464 GSPRFTHSFLQIMWFASVWVLWKKRNNRVFQNSLSDPSTFVEQVKMHSFLWLKFQQATFS 643
Query: 581 FGYHLWRQRPLACLGI 596
F YH W + PL C+G+
Sbjct: 644 FSYHDWWKHPLLCMGV 691
Score = 50.4 bits (119), Expect = 2e-06
Identities = 29/91 (31%), Positives = 45/91 (48%), Gaps = 3/91 (3%)
Frame = +3
Query: 438 TVRGVYQMLMRQERHNYDVVSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYD 497
T RG Y+ L + D W K+ P KVS++ WRL NR PTK NL++R V+
Sbjct: 117 TRRGTYRFLTISGQPLDRNQVDDVWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHVLQQT 296
Query: 498 TQFCVTGCG---QNETIDLLIIHCPIFGALW 525
C++G + ++I +FG ++
Sbjct: 297 HTACISGVAIRKRQHICFYIVIFLALFGLMF 389
>BE941032
Length = 435
Score = 65.9 bits (159), Expect = 4e-11
Identities = 32/79 (40%), Positives = 43/79 (53%)
Frame = +2
Query: 471 SIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQIKT 530
SI+ W +L NR+PTKDNL++RGVIS Q CV CG L +HC F +W +
Sbjct: 146 SIFLWCVLLNRFPTKDNLLKRGVISAIYQSCVGECGNLYDATHLFLHCNFFRQIWINVSD 325
Query: 531 WIGVFFVDPHQVLDHYYQF 549
W+ V ++ D QF
Sbjct: 326 WLSFVMVTLLRISDRLAQF 382
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 64.7 bits (156), Expect = 1e-10
Identities = 34/91 (37%), Positives = 51/91 (55%)
Frame = +1
Query: 38 QRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLL 97
++GLRQGDPLSP+LF+L A L+ L+ N G + P + +HL FADD+LL
Sbjct: 7 EKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKI-THLLFADDSLLF 183
Query: 98 GVKSWANVRALRSILVIFENMSGLKVNYNKS 128
+ + +L +++ SG VN+ KS
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKS 276
>BF650593
Length = 486
Score = 63.9 bits (154), Expect = 2e-10
Identities = 28/63 (44%), Positives = 38/63 (59%)
Frame = +3
Query: 463 HKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFG 522
HKS PLKVS WRL +N T+DNL +RGV+ ++ CV CG+ E++ CP F
Sbjct: 297 HKSVPLKVSCLVWRLFQNXLATRDNLSKRGVLDQNSIXCVXDCGREESVSHFFFECP-FS 473
Query: 523 ALW 525
+W
Sbjct: 474 XVW 482
Score = 35.4 bits (80), Expect = 0.063
Identities = 18/42 (42%), Positives = 23/42 (53%)
Frame = +1
Query: 287 GLWFRVLSSRYGEQGGRLKEGGMTGSAWWREIIKIQNGVGVE 328
GLWFR L ++YG G + S WW++I I G GVE
Sbjct: 85 GLWFRALVNKYGLNRGSITIENRGVSLWWKDICYIDFG-GVE 207
>BF520135
Length = 202
Score = 61.2 bits (147), Expect = 1e-09
Identities = 27/61 (44%), Positives = 36/61 (58%)
Frame = +3
Query: 462 WHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIF 521
W K KVSI+AWRL R PTK N+ +RG++ +D CVT C E+ L +HC +
Sbjct: 18 WRKEDLSKVSIFAWRLFHGRLPTKANVFKRGIVHHDAHMCVTRCRLIESDVHLFLHCDVL 197
Query: 522 G 522
G
Sbjct: 198 G 200
>BF643502
Length = 631
Score = 50.8 bits (120), Expect = 1e-06
Identities = 23/38 (60%), Positives = 26/38 (67%)
Frame = -1
Query: 560 LLEKVKITSLKWLKAKNVCFPFGYHLWRQRPLACLGIG 597
LL+K+K+ SL WLKAKN F FG R PL CLGIG
Sbjct: 409 LLDKIKLLSLGWLKAKNATFVFGIQQGRSNPLVCLGIG 296
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 48.9 bits (115), Expect(2) = 3e-06
Identities = 27/68 (39%), Positives = 36/68 (52%)
Frame = +2
Query: 17 CVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYS 76
CV + VLVN D RGL+QGD LSP++F++ EGL+ L+ A G S
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTS 283
Query: 77 IGMHSPTV 84
I +P V
Sbjct: 284 I*RGAPPV 307
Score = 20.4 bits (41), Expect(2) = 3e-06
Identities = 8/14 (57%), Positives = 8/14 (57%)
Frame = +1
Query: 1 MGKMGFPTLWRKWM 14
M KMGF W WM
Sbjct: 55 MIKMGFNNRWIYWM 96
>TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vigna
unguiculata}, partial (52%)
Length = 1156
Score = 49.7 bits (117), Expect = 3e-06
Identities = 23/37 (62%), Positives = 25/37 (67%)
Frame = +1
Query: 559 QLLEKVKITSLKWLKAKNVCFPFGYHLWRQRPLACLG 595
QLLEKVK+ S WLKA N FPF YH R PL C+G
Sbjct: 883 QLLEKVKLLSF*WLKATNSTFPFDYHSRRLNPLLCMG 993
>BF647389
Length = 404
Score = 48.9 bits (115), Expect = 5e-06
Identities = 40/128 (31%), Positives = 58/128 (45%)
Frame = +2
Query: 152 GKTPFMYLGLPIGGDARRLCFWDPVLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVY 211
GK P YLG+P+ +++L V +++ R+ S+ LSY G L L+K VL + Y
Sbjct: 26 GKLPSRYLGVPLS--SKKLYVIQRV-KKIICRIEN*SSKLLSYAGSLQLIKIVLFGVQPY 196
Query: 212 ALSFFRAPAGIISSIESILIKFFWGGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNV 271
F P +I I++ F G K A + IC+ K AGG V L N
Sbjct: 197 WSQVFVLP*KVIKLIQTTCRIFL*TGKSGTSKRALIAREHICLPKTAGGWNVIDLKVXNQ 376
Query: 272 ALLGKWCW 279
+ K W
Sbjct: 377 TAICKLXW 400
>BG584442
Length = 775
Score = 46.6 bits (109), Expect = 3e-05
Identities = 27/97 (27%), Positives = 48/97 (48%), Gaps = 1/97 (1%)
Frame = +1
Query: 199 ILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHRK-IAWVDWNTICMNKE 257
+++K L S+ Y +S F + IE I+ F W ++RK + W+ + ++K
Sbjct: 430 VMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGENRKGMHWMS*EKLFVHKN 609
Query: 258 AGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLS 294
GG+G FN+ +LGK L++R L+ +S
Sbjct: 610 YGGMGFTDFTTFNIPMLGKQV*SFLLNRTTLFLEKIS 720
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.140 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,193,315
Number of Sequences: 36976
Number of extensions: 401115
Number of successful extensions: 2829
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 2758
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2817
length of query: 597
length of database: 9,014,727
effective HSP length: 102
effective length of query: 495
effective length of database: 5,243,175
effective search space: 2595371625
effective search space used: 2595371625
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146549.11


