

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.3 + phase: 0
(173 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
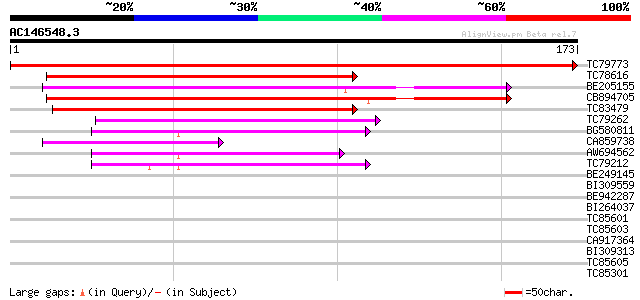
Score E
Sequences producing significant alignments: (bits) Value
TC79773 similar to PIR|C96774|C96774 AtHVA22a 65476-64429 [impo... 362 e-101
TC78616 similar to GP|11225589|gb|AAG33060.1 AtHVA22e {Arabidops... 121 1e-28
BE205155 weakly similar to PIR|C96774|C967 AtHVA22a 65476-64429... 114 3e-26
CB894705 113 3e-26
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 78 2e-15
TC79262 similar to PIR|F85433|F85433 hypothetical protein AT4g36... 67 4e-12
BG580811 similar to GP|21553813|gb unknown {Arabidopsis thaliana... 61 3e-10
CA859738 weakly similar to GP|7105620|gb|A Hypothetical protein ... 52 1e-07
AW694562 similar to GP|21553813|gb| unknown {Arabidopsis thalian... 50 6e-07
TC79212 weakly similar to GP|18652500|gb|AAL77134.1 Putative Mag... 49 1e-06
BE249145 homologue to OMNI|SO2540 response regulator {Shewanella... 30 0.49
BI309559 homologue to GP|437310|gb|A nodulin {Medicago truncatul... 30 0.64
BE942287 similar to GP|18652941|dbj protein phosphatase 2C {Arab... 29 0.83
BI264037 similar to PIR|F85433|F854 hypothetical protein AT4g367... 28 1.4
TC85601 nodulin 28 1.8
TC85603 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago t... 28 1.8
CA917364 28 2.4
BI309313 similar to GP|437310|gb|A nodulin {Medicago truncatula}... 28 2.4
TC85605 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago t... 28 2.4
TC85301 similar to GP|8096255|dbj|BAA96106.1 TrPRP2 {Trifolium r... 28 2.4
>TC79773 similar to PIR|C96774|C96774 AtHVA22a 65476-64429 [imported] -
Arabidopsis thaliana, partial (88%)
Length = 928
Score = 362 bits (930), Expect = e-101
Identities = 173/173 (100%), Positives = 173/173 (100%)
Frame = +2
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSL 60
MGGSGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSL
Sbjct: 98 MGGSGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSL 277
Query: 61 ITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYV 120
ITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYV
Sbjct: 278 ITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYV 457
Query: 121 PRKKDVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSHHTMYDETY 173
PRKKDVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSHHTMYDETY
Sbjct: 458 PRKKDVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSHHTMYDETY 616
>TC78616 similar to GP|11225589|gb|AAG33060.1 AtHVA22e {Arabidopsis
thaliana}, partial (84%)
Length = 853
Score = 121 bits (304), Expect = 1e-28
Identities = 55/95 (57%), Positives = 69/95 (71%)
Frame = +2
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKI 71
L L+ LAGPV++L+YPLYASV AIES S +DD+QWL YW++YS +TL E+
Sbjct: 170 LWTLITQLHSLAGPVVTLLYPLYASVVAIESPSKLDDEQWLAYWIIYSFLTLGEMLMQPA 349
Query: 72 LEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
LEWIPIW KL++ WLVLP F GAAY+YE +VR
Sbjct: 350 LEWIPIWYDVKLLVAAWLVLPQFMGAAYLYERFVR 454
>BE205155 weakly similar to PIR|C96774|C967 AtHVA22a 65476-64429 [imported]
- Arabidopsis thaliana, partial (24%)
Length = 610
Score = 114 bits (284), Expect = 3e-26
Identities = 59/146 (40%), Positives = 88/146 (59%), Gaps = 3/146 (2%)
Frame = +3
Query: 11 FLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAK 70
FL + + A P+++LVYP+ ASV+AIE+ S + + ++YW+L SLI LFE F
Sbjct: 3 FLKLTFKCLYHFAWPLVALVYPMCASVQAIETDSYAETKDLISYWILLSLIYLFEYAFMN 182
Query: 71 ILEWIPIWPYAKLILTCWLVLPYFTGAAYVY---EHYVRPFLANPQTINIWYVPRKKDVF 127
+L W +WPY KL++ WL++P F A+YVY +++P + N W K F
Sbjct: 183 LLLWFQLWPYTKLMIIFWLIIPDFGRASYVYNKLSRFLKPHIVTWGLNNSW-----KKWF 347
Query: 128 TKQDDIITAAEKYIKENGTEAFENLI 153
++D+ + AE Y+KENGTEA E LI
Sbjct: 348 FEKDNFLMHAEIYMKENGTEALEKLI 425
>CB894705
Length = 641
Score = 113 bits (283), Expect = 3e-26
Identities = 56/145 (38%), Positives = 91/145 (62%), Gaps = 3/145 (2%)
Frame = +1
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKI 71
L ++ ++ A P+++LVYP+ AS++AIE+ S + + ++YW+L SLI LFE F +
Sbjct: 4 LKLVFKSLHHFAWPLLALVYPMCASIQAIETDSDAEIKNIISYWILLSLIYLFEYAFMSL 183
Query: 72 LEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPF---LANPQTINIWYVPRKKDVFT 128
L W +WPY KL++ L++P F A+YVY + +RP + + + N W + F
Sbjct: 184 LLWFHLWPYIKLMIIFCLIIPDFGRASYVYNNLIRPMKLQIVSWRLNNYW-----RKCFV 348
Query: 129 KQDDIITAAEKYIKENGTEAFENLI 153
++DD + AE+Y++ENGTEA E LI
Sbjct: 349 EKDDFLMHAERYMQENGTEALEKLI 423
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein
{Plasmodium falciparum 3D7}, partial (0%)
Length = 1222
Score = 78.2 bits (191), Expect = 2e-15
Identities = 32/93 (34%), Positives = 57/93 (60%)
Frame = +2
Query: 14 VLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILE 73
+L ++ +L+ P I+L+ PLY S+R +ES +Q+ L +WVL++ + E FA +
Sbjct: 137 ILTKSLTILSWPPITLLCPLYVSIRTLESDCRSSNQRCLVFWVLFAFSMIIEREFAVLFS 316
Query: 74 WIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
W+P W + K + T L++PYF A Y+Y+ ++
Sbjct: 317 WLPWWSHVKALATILLLIPYFGAAPYIYKLLIK 415
>TC79262 similar to PIR|F85433|F85433 hypothetical protein AT4g36720
[imported] - Arabidopsis thaliana, partial (51%)
Length = 952
Score = 67.0 bits (162), Expect = 4e-12
Identities = 29/87 (33%), Positives = 49/87 (55%)
Frame = +3
Query: 27 ISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILT 86
+ + P+Y++ +AIESK+ Q+ L YW + +L E+ K++ W+P++ + K
Sbjct: 117 VGIGLPVYSTFKAIESKNQDAQQRCLVYWAAFGSFSLIEVFTDKLISWVPMYYHVKFAFL 296
Query: 87 CWLVLPYFTGAAYVYEHYVRPFLANPQ 113
WL LP GA +Y ++RPFL Q
Sbjct: 297 VWLQLPPTNGAKQLYMKHIRPFLLKHQ 377
>BG580811 similar to GP|21553813|gb unknown {Arabidopsis thaliana}, partial
(49%)
Length = 622
Score = 60.8 bits (146), Expect = 3e-10
Identities = 28/87 (32%), Positives = 44/87 (50%), Gaps = 2/87 (2%)
Frame = +2
Query: 26 VISLVYPLYASVRAIESKSPVDDQQ--WLTYWVLYSLITLFELTFAKILEWIPIWPYAKL 83
V YP Y +A+E P +Q W YW+L +L+T+ E + W+P++ KL
Sbjct: 218 VFGYAYPAYECYKAVEKNRPEIEQLRFWCQYWILVALLTVCERIGDTFISWVPMYSETKL 397
Query: 84 ILTCWLVLPYFTGAAYVYEHYVRPFLA 110
+L P G YVY+ + RP++A
Sbjct: 398 AFFIYLWYPKTKGTTYVYDSFFRPYVA 478
>CA859738 weakly similar to GP|7105620|gb|A Hypothetical protein Y71F9B.3
{Caenorhabditis elegans}, partial (26%)
Length = 428
Score = 52.0 bits (123), Expect = 1e-07
Identities = 22/55 (40%), Positives = 31/55 (56%)
Frame = +3
Query: 11 FLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFE 65
F ++ L +I VYP YAS +A+ES DD QWLTYW ++ ++L E
Sbjct: 246 FALIFFNVGGQLLSNIIGWVYPAYASFKALESSDNADDTQWLTYWTVFGFVSLVE 410
>AW694562 similar to GP|21553813|gb| unknown {Arabidopsis thaliana}, partial
(32%)
Length = 455
Score = 49.7 bits (117), Expect = 6e-07
Identities = 24/79 (30%), Positives = 38/79 (47%), Gaps = 2/79 (2%)
Frame = +2
Query: 26 VISLVYPLYASVRAIESKSPVDDQQ--WLTYWVLYSLITLFELTFAKILEWIPIWPYAKL 83
V YP Y + +E P +Q W YW+L +++T+ E + W+P++ AKL
Sbjct: 197 VFGYAYPAYECYKVVEKNKPEIEQLRFWCQYWILVAVMTVCERFGDTFVSWVPMYCEAKL 376
Query: 84 ILTCWLVLPYFTGAAYVYE 102
+L P G YVY+
Sbjct: 377 AFFIFLWYPKTKGTTYVYD 433
>TC79212 weakly similar to GP|18652500|gb|AAL77134.1 Putative Magnaporthe
grisea pathogenicity protein {Oryza sativa}, partial
(26%)
Length = 937
Score = 48.5 bits (114), Expect = 1e-06
Identities = 22/87 (25%), Positives = 46/87 (52%), Gaps = 2/87 (2%)
Frame = +3
Query: 26 VISLVYPLYASVRAIE-SKSPVDDQQ-WLTYWVLYSLITLFELTFAKILEWIPIWPYAKL 83
++ YP + + +E +K +D+ + W YW++ + T+ E ++ W+P++ KL
Sbjct: 258 LLGYAYPGFECYKTVERNKVEMDELRFWCQYWIIVAFFTVLEKFADVVIGWLPMYGELKL 437
Query: 84 ILTCWLVLPYFTGAAYVYEHYVRPFLA 110
L ++ P G YVY +RP+++
Sbjct: 438 ALFIYMWYPKTKGTGYVYNKVLRPYVS 518
>BE249145 homologue to OMNI|SO2540 response regulator {Shewanella oneidensis
MR-1}, partial (6%)
Length = 502
Score = 30.0 bits (66), Expect = 0.49
Identities = 15/44 (34%), Positives = 26/44 (59%)
Frame = -2
Query: 25 PVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTF 68
PVI + LY VR I KS + +W+ +W++ +++ F+L F
Sbjct: 225 PVIDGLRRLY--VRTIILKSEIGGLRWVVWWLVAAMLMNFDLAF 100
>BI309559 homologue to GP|437310|gb|A nodulin {Medicago truncatula}, partial
(38%)
Length = 644
Score = 29.6 bits (65), Expect = 0.64
Identities = 18/74 (24%), Positives = 35/74 (46%), Gaps = 3/74 (4%)
Frame = -1
Query: 50 QWLTY-WVLYSLITLFELTFAKILEW--IPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
+WL Y W L + + L + + ++W + +W + L WLV +F VY ++
Sbjct: 437 RWLVYRWFLNWWLVHWRLLYWRFVKWGLLNMWFVHRRFLYRWLVYRWFLNWWLVYRWFLN 258
Query: 107 PFLANPQTINIWYV 120
+L + + + W V
Sbjct: 257 WWLVHWRLLYRWLV 216
>BE942287 similar to GP|18652941|dbj protein phosphatase 2C {Arabidopsis
thaliana}, partial (21%)
Length = 527
Score = 29.3 bits (64), Expect = 0.83
Identities = 11/31 (35%), Positives = 15/31 (47%)
Frame = -1
Query: 93 YFTGAAYVYEHYVRPFLANPQTINIWYVPRK 123
+F + Y Y HY + F N Q +W RK
Sbjct: 524 FFLNSPYCYNHYTKEFQLNKQISRVWSFFRK 432
>BI264037 similar to PIR|F85433|F854 hypothetical protein AT4g36720
[imported] - Arabidopsis thaliana, partial (20%)
Length = 668
Score = 28.5 bits (62), Expect = 1.4
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = +2
Query: 27 ISLVYPLYASVRAIESKSPVDDQQWLTYW 55
+ + P+Y++ +AIESK+ Q+ L YW
Sbjct: 122 VGIGLPVYSTFKAIESKNQDAQQRCLVYW 208
>TC85601 nodulin
Length = 1933
Score = 28.1 bits (61), Expect = 1.8
Identities = 17/69 (24%), Positives = 29/69 (41%)
Frame = -3
Query: 52 LTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLAN 111
L +W+++ L ++L W W + L WLV +F VY ++ +L
Sbjct: 503 LNWWLVHRRFLYGWLVNRQLLNW---WFVYRWFLNWWLVYRWFLNWWLVYRWFLNWWLVY 333
Query: 112 PQTINIWYV 120
+N W V
Sbjct: 332 RWFLNWWLV 306
>TC85603 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago truncatula},
partial (70%)
Length = 1183
Score = 28.1 bits (61), Expect = 1.8
Identities = 16/69 (23%), Positives = 30/69 (43%)
Frame = -3
Query: 52 LTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLAN 111
L +W+++ L ++L W W + L WLV +F VY ++ +L +
Sbjct: 470 LNWWLVHRRFLYGWLVNRQLLNW---WFVYRWFLKWWLVYRWFLNWRLVYRWFINWWLVH 300
Query: 112 PQTINIWYV 120
+ + W V
Sbjct: 299 WRLLYRWLV 273
>CA917364
Length = 699
Score = 27.7 bits (60), Expect = 2.4
Identities = 16/74 (21%), Positives = 35/74 (46%), Gaps = 9/74 (12%)
Frame = +1
Query: 55 WVLYSLITLFELTFAKILEWIPIWPYAKLILTC----WLVLPYF-----TGAAYVYEHYV 105
WV S ++L ++ + ++ +W +++L C W + P A V+ H V
Sbjct: 265 WVSLSRLSLKRISHLSFMHYVGVWSLFRVLLVCPLLLWRLWPSVLILLERKIAGVHGHQV 444
Query: 106 RPFLANPQTINIWY 119
P +PQ +++++
Sbjct: 445 AP---HPQNMHVYF 477
>BI309313 similar to GP|437310|gb|A nodulin {Medicago truncatula}, partial
(20%)
Length = 542
Score = 27.7 bits (60), Expect = 2.4
Identities = 17/76 (22%), Positives = 32/76 (41%), Gaps = 7/76 (9%)
Frame = +3
Query: 52 LTYWVLYSLITLFELTFAKILEWIPI-------WPYAKLILTCWLVLPYFTGAAYVYEHY 104
L +W++Y L ++L W + W + +L WLV +F VY +
Sbjct: 279 LNWWLVYRWFLNRWLVHWRLLNWWLVHRRFLCRWLVNR*LLNWWLVYRWFLNWRLVYRWF 458
Query: 105 VRPFLANPQTINIWYV 120
+ +L + + + W V
Sbjct: 459 INWWLVHWRLLYRWLV 506
>TC85605 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago truncatula},
partial (62%)
Length = 1077
Score = 27.7 bits (60), Expect = 2.4
Identities = 19/93 (20%), Positives = 38/93 (40%), Gaps = 17/93 (18%)
Frame = -2
Query: 48 DQQWLTYWVLYSLITLFELTFAKILEWIPI-----------------WPYAKLILTCWLV 90
++Q L +W++Y + L ++L W + W + +L W V
Sbjct: 620 NRQLLNWWLMYRWFLNWWLVHWRLLYWRFVKWGLLNWWLVHRRFLYRWLVNRQLLN*WFV 441
Query: 91 LPYFTGAAYVYEHYVRPFLANPQTINIWYVPRK 123
+F VY ++ +L + + +N W V R+
Sbjct: 440 YRWFLNW*LVYRWFLNWWLVHWRLLNWWLVYRR 342
>TC85301 similar to GP|8096255|dbj|BAA96106.1 TrPRP2 {Trifolium repens},
partial (92%)
Length = 1318
Score = 27.7 bits (60), Expect = 2.4
Identities = 16/79 (20%), Positives = 35/79 (44%), Gaps = 6/79 (7%)
Frame = -1
Query: 50 QWLTYWVLYSLITLFELTFAKILEWIPIWPYA------KLILTCWLVLPYFTGAAYVYEH 103
+WL +W L+ + + + + W+ W + + +L WLV + ++Y
Sbjct: 694 RWLVHW---RLLNWWFVNWGLLNRWLVHWWFLYRWLVNRWLLNWWLVHRWLVHRWFLYRW 524
Query: 104 YVRPFLANPQTINIWYVPR 122
V +L N ++ W++ R
Sbjct: 523 LVNRWLLNWWLVHRWFLYR 467
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,144,609
Number of Sequences: 36976
Number of extensions: 89190
Number of successful extensions: 491
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 482
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 487
length of query: 173
length of database: 9,014,727
effective HSP length: 89
effective length of query: 84
effective length of database: 5,723,863
effective search space: 480804492
effective search space used: 480804492
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146548.3


