

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.9 + phase: 0
(436 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
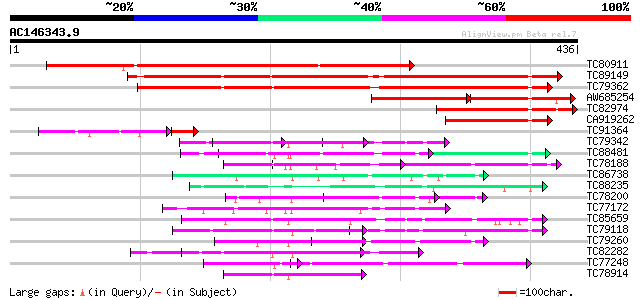
Score E
Sequences producing significant alignments: (bits) Value
TC80911 similar to GP|3668118|emb|CAA11819.1 hypothetical protei... 362 e-100
TC89149 similar to GP|20466231|gb|AAM20433.1 cell cycle switch p... 321 4e-88
TC79362 WD-repeat cell cycle regulatory protein 309 1e-84
AW685254 similar to GP|7270274|emb WD-repeat protein-like protei... 122 1e-55
TC82974 similar to GP|3668118|emb|CAA11819.1 hypothetical protei... 168 4e-42
CA919262 homologue to GP|7158292|gb| WD-repeat cell cycle regula... 99 4e-21
TC91364 similar to GP|3668118|emb|CAA11819.1 hypothetical protei... 71 3e-18
TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein h... 65 5e-11
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 63 2e-10
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 62 4e-10
TC86738 similar to PIR|T00593|T02480 sec13-related protein At2g3... 59 5e-09
TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Or... 57 1e-08
TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat prote... 54 1e-07
TC77172 similar to GP|20466472|gb|AAM20553.1 putative protein {A... 54 2e-07
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 53 3e-07
TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {... 52 6e-07
TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat pr... 50 1e-06
TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13... 50 2e-06
TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat ... 49 3e-06
TC78914 similar to GP|13937195|gb|AAK50091.1 At1g78070/F28K19_28... 49 5e-06
>TC80911 similar to GP|3668118|emb|CAA11819.1 hypothetical protein {Brassica
napus}, partial (22%)
Length = 860
Score = 362 bits (929), Expect = e-100
Identities = 188/288 (65%), Positives = 227/288 (78%), Gaps = 5/288 (1%)
Frame = +2
Query: 29 IPNRSAIDWDHATTILSTTTTNVT-KENLWLASNVYQKKLAEAADLPTRILAFRNKPRK- 86
IP RSA+D+ +A T+++ + KEN S +Y++KLA+AADLP+RILAFRNKP K
Sbjct: 2 IPCRSAMDFGYAITMVTMKNSERNRKENSSEYSVLYRQKLAQAADLPSRILAFRNKPLKP 181
Query: 87 -RNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFW 145
++ SP P P SKP R+IP+T E PD+ DDF LNLLDWG NVLSIAL++ +Y W
Sbjct: 182 IQSPSSPQPKP-SKPPRHIPQTSERKLHAPDILDDFCLNLLDWGCSNVLSIALENDVYLW 358
Query: 146 NASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHR 205
NAS+ S +E V+VDEE+GPVTSV W PDG LA+GL +S VQ+WDT ANKQL TLK GHR
Sbjct: 359 NASNKSTAELVSVDEEDGPVTSVSWCPDGSRLAIGLDSSLVQVWDTIANKQLTTLKSGHR 538
Query: 206 ARVGSLAW-NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASG 264
A V SLAW N H+LTTGGM+GKIVNNDVR+RS IN+YRGH EVCGLKWSLDGK+LASG
Sbjct: 539 AGVSSLAWNNSHILTTGGMNGKIVNNDVRVRSH-INSYRGHTDEVCGLKWSLDGKKLASG 715
Query: 265 GNDNVVHIWDMSAVSSNS-PTRWLYRFDEHKAAVKALAWCPFQGNLLA 311
G+DNVVHIWD SAVSS+S TRWL++F+EH AAVKALAWCPFQ +L+A
Sbjct: 716 GSDNVVHIWDRSAVSSSSRTTRWLHKFEEHTAAVKALAWCPFQSDLVA 859
>TC89149 similar to GP|20466231|gb|AAM20433.1 cell cycle switch protein
{Arabidopsis thaliana}, partial (71%)
Length = 1283
Score = 321 bits (822), Expect = 4e-88
Identities = 158/335 (47%), Positives = 212/335 (63%)
Frame = +2
Query: 91 SPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDS 150
+PP PPR +PKT D P L DDF LNL+DW S+N L+ L +Y W+AS+S
Sbjct: 101 TPPKPPRK-----VPKTPHKVLDAPSLQDDFYLNLVDWSSQNTLAAGLGTCVYLWSASNS 265
Query: 151 SGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGS 210
++ + +G V SV W +G +++G VQ+WD K++RT+ GGH+ R G
Sbjct: 266 KVTKLCDLGPYDG-VCSVQWTKEGSFISIGTNGGQVQIWDGTKCKKVRTM-GGHQTRTGV 439
Query: 211 LAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVV 270
LAWN +L +G D I+ +D+R+ S I GH+ EVCGLKWS D ++LASGGNDN +
Sbjct: 440 LAWNSRILASGSRDRNILQHDMRVPSDFIGKLVGHKSEVCGLKWSCDDRELASGGNDNQL 619
Query: 271 HIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGE 330
+W+ S PT R EH AAVKA+AW P Q NLL SGGG D C++ WNT G
Sbjct: 620 LVWNQH---SQQPT---LRLTEHTAAVKAIAWSPHQSNLLVSGGGTADRCIRFWNTTNGH 781
Query: 331 RMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQ 390
++NSVDTGSQVC L WSKN EL+S+HG +QNQ+ +WKYPS+ K+A L GH+ RVL++
Sbjct: 782 QLNSVDTGSQVCNLAWSKNVNELVSTHGYSQNQIMVWKYPSLAKVATLTGHSMRVLYLAM 961
Query: 391 SPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRN 425
SPDG T+ + A D+TLRFW VF + P ++
Sbjct: 962 SPDGQTIVT--GAGDETLRFWNVFPSMKTPAPVKD 1060
>TC79362 WD-repeat cell cycle regulatory protein
Length = 1800
Score = 309 bits (792), Expect = 1e-84
Identities = 149/319 (46%), Positives = 209/319 (64%)
Frame = +3
Query: 99 KPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTV 158
K R +P++ D P L DDF LNL+DW S NVL++ L + +Y WNA S ++ +
Sbjct: 603 KAPRKVPRSPYKVLDAPALQDDFYLNLVDWSSHNVLAVGLGNCVYLWNACSSKVTKLCDL 782
Query: 159 DEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVL 218
++ V SV WA G HLAVG N VQ+WD A K++R+++G HR RVG+LAW+ +L
Sbjct: 783 GVDDC-VCSVGWAQRGTHLAVGTNNGKVQIWDAARCKKIRSMEG-HRLRVGALAWSSSLL 956
Query: 219 TTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAV 278
++GG D I D+R + ++ GH+ EVCGLKWS D ++LASGGNDN + +W+ +
Sbjct: 957 SSGGRDKNIYQRDIRTQEDFVSKLSGHKSEVCGLKWSYDNRELASGGNDNKLFVWNQHS- 1133
Query: 279 SSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTG 338
T+ + ++ EH AAVKA+AW P LLASGGG D C++ WNT ++ +DTG
Sbjct: 1134-----TQPVLKYCEHTAAVKAIAWSPHLHGLLASGGGTADRCIRFWNTTTNSHLSCMDTG 1298
Query: 339 SQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVA 398
SQVC L+WSKN EL+S+HG +QNQ+ +W+YP+M K+A L GHT RVL++ SPDG T+
Sbjct: 1299SQVCNLVWSKNVNELVSTHGYSQNQIIVWRYPTMSKLATLTGHTYRVLYLAISPDGQTIV 1478
Query: 399 SAAAAADQTLRFWEVFGTP 417
+ A D+TL FW VF +P
Sbjct: 1479T--GAGDETLTFWNVFPSP 1529
>AW685254 similar to GP|7270274|emb WD-repeat protein-like protein
{Arabidopsis thaliana}, partial (35%)
Length = 657
Score = 122 bits (305), Expect(2) = 1e-55
Identities = 53/77 (68%), Positives = 65/77 (83%)
Frame = +2
Query: 279 SSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTG 338
+S SPT+WL+R ++H +AVKALAWCPFQ NLLA+GGG GD +K WNT G +NS+DTG
Sbjct: 5 TSRSPTQWLHRLEDHTSAVKALAWCPFQANLLATGGGSGDETIKFWNTHTGACLNSIDTG 184
Query: 339 SQVCALLWSKNERELLS 355
SQVC+LLW+KNERELLS
Sbjct: 185 SQVCSLLWNKNERELLS 235
Score = 113 bits (282), Expect(2) = 1e-55
Identities = 57/84 (67%), Positives = 65/84 (76%), Gaps = 3/84 (3%)
Frame = +3
Query: 355 SSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVF 414
+SHG TQNQLTLWKYPSM+KIAEL+GHTSRVLHM Q+PDG TVA+ AAAD+TLRFW F
Sbjct: 234 ASHGFTQNQLTLWKYPSMVKIAELNGHTSRVLHMAQNPDGCTVAT--AAADETLRFWNAF 407
Query: 415 GTPPA---APPKRNKMPFADSNRI 435
GTP A PK PF+ +RI
Sbjct: 408 GTPEVATKAAPKARAEPFSHVSRI 479
>TC82974 similar to GP|3668118|emb|CAA11819.1 hypothetical protein {Brassica
napus}, partial (19%)
Length = 569
Score = 168 bits (425), Expect = 4e-42
Identities = 83/108 (76%), Positives = 96/108 (88%)
Frame = +3
Query: 329 GERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHM 388
G R++SVDTGS+VCALLW+KNERELLSSHGLT+NQ+TLWKYPSM+K+AEL+GHTSRVL+M
Sbjct: 6 GARLDSVDTGSEVCALLWNKNERELLSSHGLTKNQITLWKYPSMVKMAELNGHTSRVLYM 185
Query: 389 TQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRNKMPFADSNRIR 436
TQSPDG TVA+ AAAD+TLRFW VFGTP A PK N PFA+ NRIR
Sbjct: 186 TQSPDGCTVAT--AAADETLRFWNVFGTPKAT-PKTNHEPFANFNRIR 320
>CA919262 homologue to GP|7158292|gb| WD-repeat cell cycle regulatory protein
{Medicago truncatula}, partial (23%)
Length = 820
Score = 98.6 bits (244), Expect = 4e-21
Identities = 45/82 (54%), Positives = 61/82 (73%)
Frame = -2
Query: 336 DTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGS 395
DTGSQVC L+ KN +L+S+HG +QNQ+ +W+YP+M K+A L GHT RVL++ SPDG
Sbjct: 786 DTGSQVCNLVCPKNVNDLVSTHGYSQNQIIVWRYPTMSKLATLPGHTYRVLYLAISPDGQ 607
Query: 396 TVASAAAAADQTLRFWEVFGTP 417
T+ + A D+TLRFW VF +P
Sbjct: 606 TIVT--GAGDETLRFWNVFPSP 547
>TC91364 similar to GP|3668118|emb|CAA11819.1 hypothetical protein {Brassica
napus}, partial (25%)
Length = 679
Score = 71.2 bits (173), Expect(2) = 3e-18
Identities = 51/115 (44%), Positives = 67/115 (57%), Gaps = 13/115 (11%)
Frame = +2
Query: 23 QNIDRFIPNRSAIDWDHATTILSTTTTNVTKENLWLAS---NVYQKKLAEAADLP-TRIL 78
+N+DRFIPNRSA+D+D+A +++ KEN + S Y+K LAE+ ++ TRIL
Sbjct: 281 ENLDRFIPNRSAMDFDYAHYMVTEGAKG--KENPEVCSPSREAYRKLLAESLNMNRTRIL 454
Query: 79 AFRNKPRKRNVISPPPPPRS---------KPMRYIPKTCEGTFDLPDLSDDFSLN 124
AF+NKP V S P S KP R IP+T E T D PDL DD+ LN
Sbjct: 455 AFKNKP-PTPVDSIPHELTSSSLQEDKTIKPRRIIPQTSERTLDAPDLVDDYYLN 616
Score = 38.1 bits (87), Expect(2) = 3e-18
Identities = 15/21 (71%), Positives = 18/21 (85%)
Frame = +1
Query: 125 LLDWGSRNVLSIALDHTIYFW 145
LLDWGS NVL+IAL +T+Y W
Sbjct: 616 LLDWGSANVLAIALGNTVYLW 678
>TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein homolog
{Arabidopsis thaliana}, partial (47%)
Length = 873
Score = 65.1 bits (157), Expect = 5e-11
Identities = 38/127 (29%), Positives = 62/127 (47%), Gaps = 7/127 (5%)
Frame = +3
Query: 157 TVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW--N 214
T+ V SV ++PDGR LA G ++ V+ WD + T GH+ V +AW +
Sbjct: 480 TISGHGEAVLSVAFSPDGRQLASGSGDTTVRFWDLGTQTPMYTCT-GHKNWVLCIAWSPD 656
Query: 215 GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKW-----SLDGKQLASGGNDNV 269
G L +G M G+++ D + Q+ N GH++ + G+ W + ++ S D
Sbjct: 657 GKYLVSGSMSGELICWDPQTGKQLGNALTGHKKWITGISWEPVHLNAPCRRFVSASKDGD 836
Query: 270 VHIWDMS 276
IWD+S
Sbjct: 837 ARIWDVS 857
Score = 52.4 bits (124), Expect = 3e-07
Identities = 32/98 (32%), Positives = 46/98 (46%)
Frame = +3
Query: 241 TYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKAL 300
T GH V + +S DG+QLASG D V WD+ + +Y HK V +
Sbjct: 480 TISGHGEAVLSVAFSPDGRQLASGSGDTTVRFWDLGTQTP------MYTCTGHKNWVLCI 641
Query: 301 AWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTG 338
AW P G L SG G+ + W+ G+++ + TG
Sbjct: 642 AWSP-DGKYLVSGSMSGE--LICWDPQTGKQLGNALTG 746
Score = 44.7 bits (104), Expect = 7e-05
Identities = 22/83 (26%), Positives = 38/83 (45%)
Frame = +3
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
R + S + D T+ FW+ + T + V + W+PDG++L G + + WD
Sbjct: 534 RQLASGSGDTTVRFWDLGTQT--PMYTCTGHKNWVLCIAWSPDGKYLVSGSMSGELICWD 707
Query: 191 TAANKQLRTLKGGHRARVGSLAW 213
KQL GH+ + ++W
Sbjct: 708 PQTGKQLGNALTGHKKWITGISW 776
Score = 33.1 bits (74), Expect = 0.22
Identities = 16/38 (42%), Positives = 23/38 (60%)
Frame = +3
Query: 376 AELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
A + GH VL + SPDG +AS + D T+RFW++
Sbjct: 477 ATISGHGEAVLSVAFSPDGRQLAS--GSGDTTVRFWDL 584
Score = 28.9 bits (63), Expect = 4.1
Identities = 18/68 (26%), Positives = 32/68 (46%)
Frame = +3
Query: 338 GSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTV 397
G V ++ +S + R+L S G T + W + + GH + VL + SPDG +
Sbjct: 495 GEAVLSVAFSPDGRQLASGSGDTT--VRFWDLGTQTPMYTCTGHKNWVLCIAWSPDGKYL 668
Query: 398 ASAAAAAD 405
S + + +
Sbjct: 669 VSGSMSGE 692
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 63.2 bits (152), Expect = 2e-10
Identities = 47/197 (23%), Positives = 86/197 (42%), Gaps = 2/197 (1%)
Frame = +1
Query: 132 NVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDT 191
++ + + D T +WN G+ T + + + P G++L + +LWD
Sbjct: 1120 HLATASADRTAKYWN---DQGALLGTFKGHLERLARIAFHPSGKYLGTASYDKTWRLWDV 1290
Query: 192 AANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREV 249
++L L+ GH V L ++ G + + G+D D+R ++ GH + +
Sbjct: 1291 ETEEEL-LLQEGHSRSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVL-ALEGHVKSI 1464
Query: 250 CGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNL 309
G+ +S +G LA+GG DN IWD+ S LY H + + + P +G
Sbjct: 1465 LGISFSPNGYHLATGGEDNTCRIWDLRKKKS------LYTIPAHSNLISQVKFEPQEGYF 1626
Query: 310 LASGGGGGDCCVKLWNT 326
L + D K+W++
Sbjct: 1627 LVT--ASYDMTAKVWSS 1671
Score = 55.5 bits (132), Expect = 4e-08
Identities = 59/299 (19%), Positives = 110/299 (36%), Gaps = 43/299 (14%)
Frame = +1
Query: 161 EEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKG------------------ 202
++ P+T ++ DG+ LA +LW K++ TLKG
Sbjct: 946 DDRPLTGCSFSRDGKGLATCSFTGATKLWSMPNVKKVSTLKGHTQRATDVAYSPVHKNHL 1125
Query: 203 -----------------------GHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQ 237
GH R+ +A+ +G L T D DV +
Sbjct: 1126 ATASADRTAKYWNDQGALLGTFKGHLERLARIAFHPSGKYLGTASYDKTWRLWDVETEEE 1305
Query: 238 IINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAV 297
++ GH R V GL + DG AS G D + +WD+ R + + H ++
Sbjct: 1306 LL-LQEGHSRSVYGLDFHHDGSLAASCGLDALARVWDLRT------GRSVLALEGHVKSI 1464
Query: 298 KALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSH 357
+++ P G LA+GG C ++W+ + + ++ S + + + + +
Sbjct: 1465 LGISFSP-NGYHLATGGEDNTC--RIWDLRKKKSLYTIPAHSNLISQVKFEPQEGYFLVT 1635
Query: 358 GLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGT 416
+W + L GH ++V + DG + + + D+T++ W T
Sbjct: 1636 ASYDMTAKVWSSRDFKPVKTLSGHEAKVTSLDVLGDGGYIVT--VSHDRTIKLWSXXTT 1806
Score = 40.0 bits (92), Expect = 0.002
Identities = 31/153 (20%), Positives = 63/153 (40%), Gaps = 5/153 (3%)
Frame = +1
Query: 135 SIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAAN 194
S LD W+ +G + ++ + + ++P+G HLA G ++ ++WD
Sbjct: 1378 SCGLDALARVWDLR--TGRSVLALEGHVKSILGISFSPNGYHLATGGEDNTCRIWDLRKK 1551
Query: 195 KQLRTLKGGHRARVGSLAWN---GHVLTTGGMD--GKIVNNDVRLRSQIINTYRGHRREV 249
K L T+ H + + + G+ L T D K+ ++ + + T GH +V
Sbjct: 1552 KSLYTIPA-HSNLISQVKFEPQEGYFLVTASYDMTAKVWSSR---DFKPVKTLSGHEAKV 1719
Query: 250 CGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNS 282
L DG + + +D + +W ++
Sbjct: 1720 TSLDVLGDGGYIVTVSHDRTIKLWSXXTTXXHA 1818
Score = 35.8 bits (81), Expect = 0.033
Identities = 48/210 (22%), Positives = 74/210 (34%), Gaps = 40/210 (19%)
Frame = +1
Query: 244 GHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWC 303
G R + G +S DGK LA+ +W M V S + H +A+
Sbjct: 943 GDDRPLTGCSFSRDGKGLATCSFTGATKLWSMPNVKKVSTLK------GHTQRATDVAYS 1104
Query: 304 PFQGNLLASGGGGGDCCVKLWN---------TGMGERMNSV---DTGSQVCAL------- 344
P N LA+ D K WN G ER+ + +G +
Sbjct: 1105 PVHKNHLAT--ASADRTAKYWNDQGALLGTFKGHLERLARIAFHPSGKYLGTASYDKTWR 1278
Query: 345 LWS-KNERELLSSHGLTQNQLTL--------------------WKYPSMLKIAELHGHTS 383
LW + E ELL G +++ L W + + L GH
Sbjct: 1279 LWDVETEEELLLQEGHSRSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVLALEGHVK 1458
Query: 384 RVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
+L ++ SP+G +A+ D T R W++
Sbjct: 1459 SILGISFSPNGYHLAT--GGEDNTCRIWDL 1542
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 62.0 bits (149), Expect = 4e-10
Identities = 54/224 (24%), Positives = 92/224 (40%), Gaps = 2/224 (0%)
Frame = +1
Query: 203 GHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQ 260
GH++ V ++ +N G V+ +G D +I +V + +GH+ V L W+ DG Q
Sbjct: 307 GHQSAVYTMKFNPTGSVVASGSHDKEIFLWNVHGDCKNFMVLKGHKNAVLDLHWTSDGTQ 486
Query: 261 LASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCC 320
+ S D + +WD + + + EH + V + CP + G D
Sbjct: 487 IISASPDKTLRLWD------TETGKQIKKMVEHLSYVNSC--CPTRRGPPLVVSGSDDGT 642
Query: 321 VKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHG 380
KLW+ + + Q+ A+ +S ++ + G N + +W L G
Sbjct: 643 AKLWDMRQRGSIQTFPDKYQITAVSFSDASDKIYT--GGIDNDVKIWDLRKGEVTMTLQG 816
Query: 381 HTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKR 424
H + M SPDGS + + D L W++ P AP R
Sbjct: 817 HQDMITSMQLSPDGSYLLT--NGMDCKLCIWDM---RPYAPQNR 933
Score = 55.8 bits (133), Expect = 3e-08
Identities = 41/151 (27%), Positives = 74/151 (48%), Gaps = 11/151 (7%)
Frame = +1
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGS--LAWNGHVLTTGG 222
+T+V ++ + G ++ V++WD + TL+G H+ + S L+ +G L T G
Sbjct: 703 ITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQG-HQDMITSMQLSPDGSYLLTNG 879
Query: 223 MDGKIVNNDVRL---RSQIINTYRGHRREV------CGLKWSLDGKQLASGGNDNVVHIW 273
MD K+ D+R +++ + GH+ CG WS DG ++ +G +D +V+IW
Sbjct: 880 MDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCG--WSPDGSKVTAGSSDRMVYIW 1053
Query: 274 DMSAVSSNSPTRWLYRFDEHKAAVKALAWCP 304
D ++ R LY+ H +V + P
Sbjct: 1054DTTS------RRILYKLPGHNGSVNECVFHP 1128
Score = 27.7 bits (60), Expect = 9.1
Identities = 16/38 (42%), Positives = 20/38 (52%)
Frame = +1
Query: 378 LHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFG 415
L GH S V M +P GS VAS + D+ + W V G
Sbjct: 301 LTGHQSAVYTMKFNPTGSVVAS--GSHDKEIFLWNVHG 408
>TC86738 similar to PIR|T00593|T02480 sec13-related protein At2g30050 -
Arabidopsis thaliana, complete
Length = 1278
Score = 58.5 bits (140), Expect = 5e-09
Identities = 72/276 (26%), Positives = 109/276 (39%), Gaps = 33/276 (11%)
Frame = +2
Query: 126 LDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPD--GRHLAVGLTN 183
+D+ + + + + DHTI S+S+ T+ +GPV V WA G LA +
Sbjct: 176 MDYYGKRLATASSDHTIKIIGVSNSASQHLATLTGHQGPVWEVAWAHPKFGSLLASCSYD 355
Query: 184 SHVQLW-DTAANKQLRT-LKGGHRARVGSLAWNGH----VLTTGGMDGKIVNNDVRLRS- 236
V LW + N+ ++ + H++ V S+AW H L DG I R
Sbjct: 356 GRVILWKEGNQNEWIQAHVFDEHKSSVNSVAWAPHELGLCLACASSDGNISVFTARADGG 535
Query: 237 -QIINTYRGHRREVCGLKWS-------------LDGKQ-LASGGNDNVVHIWDMSAVSSN 281
+ H V + W+ LD Q L SGG DN V +W +S +
Sbjct: 536 WDTSRIDQAHPVGVTSVSWAPSMAPGALVGSGLLDPVQKLCSGGCDNTVKVWKLS--NGQ 709
Query: 282 SPTRWLYRFDEHKAAVKALAWCPFQG---NLLASGGGGGDCCVKLWNTGM------GERM 332
+H V+ +AW P G + +AS G V +W G G+ +
Sbjct: 710 WKMDCFPALQKHNDWVRDVAWAPNLGLPKSTIASASQDGK--VIIWTAGKEGDHWEGKDL 883
Query: 333 NSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWK 368
N D + V + WS L + G N +TLWK
Sbjct: 884 N--DFKTPVWRVSWSLTGNILAVADG--NNNVTLWK 979
Score = 29.6 bits (65), Expect = 2.4
Identities = 18/63 (28%), Positives = 29/63 (45%), Gaps = 1/63 (1%)
Frame = +2
Query: 133 VLSIALDHTIYFWNASDSSGS-EFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDT 191
+ S + D + W A E +++ + PV V W+ G LAV N++V LW
Sbjct: 803 IASASQDGKVIIWTAGKEGDHWEGKDLNDFKTPVWRVSWSLTGNILAVADGNNNVTLWKE 982
Query: 192 AAN 194
A +
Sbjct: 983 AVD 991
>TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Oryza
sativa}, partial (91%)
Length = 1080
Score = 57.4 bits (137), Expect = 1e-08
Identities = 65/284 (22%), Positives = 105/284 (36%), Gaps = 9/284 (3%)
Frame = +2
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
DHTI FW A SG + T+ + V + PD R+LA N H++L+D +N
Sbjct: 251 DHTIRFWEA--KSGRCYRTIQYPDSQVNRLEITPDKRYLAAA-GNPHIRLFDVNSNSPQP 421
Query: 199 TLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDG 258
++++GH V + + DG
Sbjct: 422 V-----------MSYDGHT-----------------------------SNVMAVGFQCDG 481
Query: 259 KQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGD 318
+ SG D V IWD+ A R E +AAV + P Q L++ G
Sbjct: 482 NWMYSGSEDGTVKIWDLRAPGCQ-------REYESRAAVNTVVLHPNQTELISGDQNGN- 637
Query: 319 CCVKLW----NTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLK 374
+++W N+ E + VDT + ++W + +++G L +M
Sbjct: 638 --IRVWDLTANSCSCELVPEVDTAVRSLTVMWDGSLVVAANNNGTCYVWRLLRGTQTMTN 811
Query: 375 IAELH---GHTSRVLHMTQSPDGSTVAS--AAAAADQTLRFWEV 413
LH H +L SP+ A A++D T++ W V
Sbjct: 812 FEPLHKLQAHNGYILKCVLSPEFCDPHRYLATASSDHTVKIWNV 943
Score = 28.9 bits (63), Expect = 4.1
Identities = 15/61 (24%), Positives = 30/61 (48%)
Frame = +2
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
R + + + DHT+ WN D + + T+ + + ++ DG +L ++S +LW
Sbjct: 893 RYLATASSDHTVKIWNVDDFTLEK--TLIGHQRRLWDCVFSVDGAYLITASSDSTARLWS 1066
Query: 191 T 191
T
Sbjct: 1067T 1069
>TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana, partial (80%)
Length = 1824
Score = 53.9 bits (128), Expect = 1e-07
Identities = 46/173 (26%), Positives = 79/173 (45%), Gaps = 9/173 (5%)
Frame = +1
Query: 167 SVCWAP--DGRHLAVGLTNSHVQLWD--TAANKQL-RTLKGGHRARVGSLAWNG---HVL 218
++ W+P GR L G N+ + LW+ +AA + +T GH V L W+ HV
Sbjct: 721 AIDWSPLVPGR-LVSGDCNNSIYLWEPTSAATWNIEKTPFTGHTDSVEDLQWSPTEPHVF 897
Query: 219 TTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWS-LDGKQLASGGNDNVVHIWDMSA 277
+ +D I D RL +++ H+ +V L W+ L LASG +D + I D+
Sbjct: 898 ASCSVDKSIAIWDTRLGRSPAASFQAHKADVNVLSWNRLASCMLASGSDDGTISIRDLRL 1077
Query: 278 VSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGE 330
+ + F+ HK + ++ W P + + LA D + +W+ G+
Sbjct: 1078LKEGDSV--VAHFEYHKHPITSIEWSPHEASSLAV--SSADNQLTIWDLFFGK 1224
Score = 41.2 bits (95), Expect = 8e-04
Identities = 36/130 (27%), Positives = 53/130 (40%), Gaps = 4/130 (3%)
Frame = +1
Query: 242 YRGHRREVCGLKWS-LDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKAL 300
+ GH V L+WS + AS D + IWD S + + F HKA V L
Sbjct: 835 FTGHTDSVEDLQWSPTEPHVFASCSVDKSIAIWDTRLGRSPAAS-----FQAHKADVNVL 999
Query: 301 AWCPFQGNLLASGGGGGDCCV---KLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSH 357
+W +LASG G + +L G + + ++ WS +E L+
Sbjct: 1000SWNRLASCMLASGSDDGTISIRDLRLLKEGDSVVAHFEYHKHPITSIEWSPHEASSLAVS 1179
Query: 358 GLTQNQLTLW 367
NQLT+W
Sbjct: 1180S-ADNQLTIW 1206
>TC77172 similar to GP|20466472|gb|AAM20553.1 putative protein {Arabidopsis
thaliana}, partial (78%)
Length = 4130
Score = 53.5 bits (127), Expect = 2e-07
Identities = 64/244 (26%), Positives = 106/244 (43%), Gaps = 22/244 (9%)
Frame = +3
Query: 118 SDDFSLNLLDWGSRNVLSIALDHTIYFWNA-----SDSSGSEFVT-VDEEEGPVTSVCW- 170
S++F+L L+ G +D I WN S+ + S V + +GPV + +
Sbjct: 345 SEEFALGLVAGG-------LVDGNIDLWNPLSLIRSEENESSLVGHLVRHKGPVRGLEFN 503
Query: 171 --APDGRHLAVGLTNSHVQLWDTAANKQ---LRTLKGGHRARVGS---LAWNG---HVLT 219
AP+ LA G + + +WD A + LKG A G L+WN H+L
Sbjct: 504 SIAPN--LLASGAEDGEICIWDLANPLEPTHFPPLKGSGSASQGEVSFLSWNSKVQHILA 677
Query: 220 TGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLD-GKQLASGGNDN---VVHIWDM 275
+ +G V D++ + +I+ RR L+W D QLA +++ + +WDM
Sbjct: 678 STSYNGTTVVWDLKKQKSVISVVDPVRRRGSALQWHPDVATQLAVASDEDGSPSIKLWDM 857
Query: 276 SAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSV 335
++ +P + F H V A++WCP + L + G D W+T GE +
Sbjct: 858 R--NTMTPVK---EFVGHSRGVIAMSWCPNDSSYLLT--CGKDSRTICWDTISGEIAYEL 1016
Query: 336 DTGS 339
G+
Sbjct: 1017PAGT 1028
Score = 30.8 bits (68), Expect = 1.1
Identities = 52/266 (19%), Positives = 104/266 (38%), Gaps = 42/266 (15%)
Frame = +3
Query: 193 ANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIV-------------------NNDVR 233
++ Q+ +KG +R+ + +LA + L G M G + + ++
Sbjct: 99 SDSQMACIKGVNRSALVALAPDAPYLAAGTMAGAVDLLFSSSANLEIFKIDFQSDDPELP 278
Query: 234 LRSQIINTYRGHRREVCGLKWSLDGKQ--------LASGGNDNVVHIWDMSAV--SSNSP 283
L ++ ++ R +R L W +G +A G D + +W+ ++ S +
Sbjct: 279 LVAEYPSSDRFNR-----LSWGRNGSSSEEFALGLVAGGLVDGNIDLWNPLSLIRSEENE 443
Query: 284 TRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMG---------ERMNS 334
+ + HK V+ L + NLLASG G+ C+ W+ + S
Sbjct: 444 SSLVGHLVRHKGPVRGLEFNSIAPNLLASGAEDGEICI--WDLANPLEPTHFPPLKGSGS 617
Query: 335 VDTGSQVCALLWSKNERELLSSHGLTQNQLT--LWKYPSMLKIAELHGHTSRVLHMTQSP 392
G +V L W+ + +L+S + L K S++ + + L P
Sbjct: 618 ASQG-EVSFLSWNSKVQHILASTSYNGTTVVWDLKKQKSVISVVDPVRRRGSALQW--HP 788
Query: 393 DGSTVASAAAAAD--QTLRFWEVFGT 416
D +T + A+ D +++ W++ T
Sbjct: 789 DVATQLAVASDEDGSPSIKLWDMRNT 866
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 52.8 bits (125), Expect = 3e-07
Identities = 75/307 (24%), Positives = 128/307 (41%), Gaps = 26/307 (8%)
Frame = +2
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGP---VTSVCWAPDGRHLAVGLTNSHVQLW 189
+++ + D +I W+ + + V G V V + DG+ G + ++LW
Sbjct: 152 IVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDGELRLW 331
Query: 190 DTAANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKI-VNNDVRLRSQIINTYRGHR 246
D A R GH V S+A+ + + + D I + N + I H
Sbjct: 332 DLNAGTSARRFV-GHTKDVLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDAHS 508
Query: 247 REVCGLKWSLDGKQ--LASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCP 304
V +++S Q + S D V +W+++ + H V +A P
Sbjct: 509 DWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKLRN------TLAGHSGYVNTVAVSP 670
Query: 305 FQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQL 364
G+L ASGG G + LW+ G+R+ S+D GS + AL +S N L ++ T++ +
Sbjct: 671 -DGSLCASGGKDG--VILLWDLAEGKRLYSLDAGSIIHALCFSPNRYWLCAA---TESSI 832
Query: 365 TLWKYPSM-----LKI-------AELHGHTS---RVLHMTQ---SPDGSTVASAAAAADQ 406
+W S LK+ A + G T+ +V++ T S DGST+ S D
Sbjct: 833 KIWDLESKSIVEDLKVDLKTEADAAIGGDTTTKKKVIYCTSLNWSADGSTLFS--GYTDG 1006
Query: 407 TLRFWEV 413
+R W +
Sbjct: 1007VVRVWGI 1027
>TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {Arabidopsis
thaliana}, partial (44%)
Length = 1098
Score = 51.6 bits (122), Expect = 6e-07
Identities = 38/162 (23%), Positives = 72/162 (43%), Gaps = 12/162 (7%)
Frame = +2
Query: 126 LDWGSR-NVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNS 184
+DW + + S + D+ IY ++ ++ T +G V V W P G LA +
Sbjct: 212 VDWRTNTSFASSSTDNMIYVCKIGENHPTQ--TFAGHQGEVNCVKWDPTGTLLASCSDDI 385
Query: 185 HVQLWDTAANKQLRTLKGGHRARVGSLAWNGH-----------VLTTGGMDGKIVNNDVR 233
++W +K L + H + ++ W+ +L + D + D+
Sbjct: 386 TAKIWSVKQDKYLHDFRE-HSKEIYTIRWSPTGPGTNNPNKKLLLASASFDSTVKLWDIE 562
Query: 234 LRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM 275
L ++I++ GHR V + +S +G+ +ASG D +HIW +
Sbjct: 563 L-GKLIHSLNGHRHPVYSVAFSPNGEYIASGSLDKSLHIWSL 685
Score = 47.4 bits (111), Expect = 1e-05
Identities = 39/160 (24%), Positives = 69/160 (42%), Gaps = 8/160 (5%)
Frame = +2
Query: 262 ASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCV 321
AS DN++++ + N PT+ F H+ V + W P G LLAS D
Sbjct: 239 ASSSTDNMIYV---CKIGENHPTQ---TFAGHQGEVNCVKWDP-TGTLLASCSD--DITA 391
Query: 322 KLWNTGMGERMNSV-DTGSQVCALLWSKN-------ERELLSSHGLTQNQLTLWKYPSML 373
K+W+ + ++ + ++ + WS ++LL + + + LW
Sbjct: 392 KIWSVKQDKYLHDFREHSKEIYTIRWSPTGPGTNNPNKKLLLASASFDSTVKLWDIELGK 571
Query: 374 KIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
I L+GH V + SP+G +AS + D++L W +
Sbjct: 572 LIHSLNGHRHPVYSVAFSPNGEYIAS--GSLDKSLHIWSL 685
Score = 37.4 bits (85), Expect = 0.011
Identities = 20/80 (25%), Positives = 37/80 (46%)
Frame = +2
Query: 135 SIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAAN 194
S + D T+ W+ G +++ PV SV ++P+G ++A G + + +W
Sbjct: 521 SASFDSTVKLWDIE--LGKLIHSLNGHRHPVYSVAFSPNGEYIASGSLDKSLHIWSLKEG 694
Query: 195 KQLRTLKGGHRARVGSLAWN 214
K +RT G + + WN
Sbjct: 695 KIIRTYNGS--GGIFEVCWN 748
>TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-like
{Arabidopsis thaliana}, partial (50%)
Length = 1227
Score = 50.4 bits (119), Expect = 1e-06
Identities = 34/137 (24%), Positives = 65/137 (46%), Gaps = 1/137 (0%)
Frame = +2
Query: 233 RLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDE 292
++ S+ + H EV +++S DGK LA+ ND IW+ V +N ++
Sbjct: 506 QIPSRTLQILDAHDDEVWFVQFSHDGKYLATASNDRTAIIWE---VDTNDGLSMKHKLSG 676
Query: 293 HKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVD-TGSQVCALLWSKNER 351
H+ +V +++W P LL G + V+ W+ G+ + + GS + + W +
Sbjct: 677 HQKSVSSVSWSPNGQELLTC---GVEEAVRRWDVSTGKCLQVYEKNGSGLISCAWFPCGK 847
Query: 352 ELLSSHGLTQNQLTLWK 368
+LS GL+ + +W+
Sbjct: 848 YILS--GLSDKSICMWE 892
Score = 44.3 bits (103), Expect = 9e-05
Identities = 30/122 (24%), Positives = 56/122 (45%), Gaps = 4/122 (3%)
Frame = +2
Query: 158 VDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLW--DTAANKQLRTLKGGHRARVGSLAW-- 213
+D + V V ++ DG++LA + +W DT ++ GH+ V S++W
Sbjct: 533 LDAHDDEVWFVQFSHDGKYLATASNDRTAIIWEVDTNDGLSMKHKLSGHQKSVSSVSWSP 712
Query: 214 NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIW 273
NG L T G++ + DV + + Y + + W GK + SG +D + +W
Sbjct: 713 NGQELLTCGVEEAVRRWDVS-TGKCLQVYEKNGSGLISCAWFPCGKYILSGLSDKSICMW 889
Query: 274 DM 275
++
Sbjct: 890 EL 895
Score = 35.0 bits (79), Expect = 0.057
Identities = 23/80 (28%), Positives = 40/80 (49%), Gaps = 1/80 (1%)
Frame = +2
Query: 132 NVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDT 191
++LSI ++ I +N ++ F+ EE+ +TS + D R L V L N + LW+
Sbjct: 974 HILSICKENAILLFN-KETKVERFI---EEDQTITSFSLSKDSRFLLVNLLNQEIHLWNI 1141
Query: 192 AAN-KQLRTLKGGHRARVGS 210
+ K + K R+R G+
Sbjct: 1142 EGDLKLVGKYKSHRRSRFGN 1201
Score = 34.7 bits (78), Expect = 0.074
Identities = 30/140 (21%), Positives = 62/140 (43%), Gaps = 4/140 (2%)
Frame = +2
Query: 139 DHTIYFWNASDSSGSEFV-TVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQL 197
D T W + G + + V+SV W+P+G+ L V+ WD + K L
Sbjct: 608 DRTAIIWEVDTNDGLSMKHKLSGHQKSVSSVSWSPNGQELLTCGVEEAVRRWDVSTGKCL 787
Query: 198 RTLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRR-EVCGLKW 254
+ + + S AW G + +G D I + L + + +++G + ++ L+
Sbjct: 788 QVYEKNGSGLI-SCAWFPCGKYILSGLSDKSICMWE--LDGKEVESWKGQKTLKISDLEI 958
Query: 255 SLDGKQLASGGNDNVVHIWD 274
+ DG+ + S +N + +++
Sbjct: 959 TGDGEHILSICKENAILLFN 1018
>TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13.70 -
Arabidopsis thaliana, partial (84%)
Length = 904
Score = 49.7 bits (117), Expect = 2e-06
Identities = 49/183 (26%), Positives = 78/183 (41%), Gaps = 3/183 (1%)
Frame = +2
Query: 94 PPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGS 153
PP MR+ PK G S+NL D + ++ + SD SGS
Sbjct: 377 PPSEVWQMRFDPK---GAILAVAGGGSASVNLWDTSTWELVVTLSIPRVEGPKPSDKSGS 547
Query: 154 EFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW 213
+ V SV W+PDG+ LA G + + ++D K L L+ GH V SL +
Sbjct: 548 KKF--------VLSVAWSPDGKRLACGSMDGTISVFDVQRAKFLHHLE-GHFMPVRSLVY 700
Query: 214 NGH---VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVV 270
+ + +L + DG + D ++ + T H V + S +G +A+G +D V
Sbjct: 701 SPYDPRLLFSASDDGNVHMYDAEGKA-LXGTMSXHASWVLCVDVSPNGAAIATGSSDKTV 877
Query: 271 HIW 273
+W
Sbjct: 878 RLW 886
Score = 49.3 bits (116), Expect = 3e-06
Identities = 48/200 (24%), Positives = 84/200 (42%), Gaps = 14/200 (7%)
Frame = +2
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
+L+ +LD T+ W SD E G V SV P G A +S V+++D
Sbjct: 173 LLTGSLDETVRLWK-SDDLVLERTNTGHCLG-VASVAAHPLGSIAASSSLDSFVRVFDVD 346
Query: 193 ANKQLRTL--------------KGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQI 238
+N + TL KG A G + + ++ T + + + R+
Sbjct: 347 SNATIATLEAPPSEVWQMRFDPKGAILAVAGGGSASVNLWDTSTWELVVTLSIPRVEGPK 526
Query: 239 INTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVK 298
+ G ++ V + WS DGK+LA G D + ++D+ ++L+ + H V+
Sbjct: 527 PSDKSGSKKFVLSVAWSPDGKRLACGSMDGTISVFDVQR------AKFLHHLEGHFMPVR 688
Query: 299 ALAWCPFQGNLLASGGGGGD 318
+L + P+ LL S G+
Sbjct: 689 SLVYSPYDPRLLFSASDDGN 748
Score = 32.0 bits (71), Expect = 0.48
Identities = 61/285 (21%), Positives = 105/285 (36%), Gaps = 70/285 (24%)
Frame = +2
Query: 197 LRTLKGGHRARVGSLAW------NGHVLTTGGMDGKI---VNNDVRLRSQIINTYRGHRR 247
++++ H V ++ W +L TG +D + ++D+ L T GH
Sbjct: 92 IKSIDNAHDDSVWAVTWAPATATRPPLLLTGSLDETVRLWKSDDLVLE----RTNTGHCL 259
Query: 248 EVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTR------WLYRFDEHKAAVKALA 301
V + G AS D+ V ++D+ + ++ + W RFD K A+ A+A
Sbjct: 260 GVASVAAHPLGSIAASSSLDSFVRVFDVDSNATIATLEAPPSEVWQMRFDP-KGAILAVA 436
Query: 302 WCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVD------------TGSQ--VCALLWS 347
GGG V LW+T E + ++ +GS+ V ++ WS
Sbjct: 437 -------------GGGSASVNLWDTSTWELVVTLSIPRVEGPKPSDKSGSKKFVLSVAWS 577
Query: 348 KNEREL----------------------LSSHGLTQNQLTLWKY-PSMLKIAELHG---- 380
+ + L L H + L Y P +L A G
Sbjct: 578 PDGKRLACGSMDGTISVFDVQRAKFLHHLEGHFMPVRSLVYSPYDPRLLFSASDDGNVHM 757
Query: 381 --------------HTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
H S VL + SP+G+ +A+ ++D+T+R W
Sbjct: 758 YDAEGKALXGTMSXHASWVLCVDVSPNGAAIAT--GSSDKTVRLW 886
>TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat containing
protein {Oryza sativa}, partial (92%)
Length = 2005
Score = 49.3 bits (116), Expect = 3e-06
Identities = 42/185 (22%), Positives = 79/185 (42%)
Frame = +1
Query: 217 VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMS 276
++ TG +D V D R Q++ T GH ++V +K+ G+ + +G D V +W S
Sbjct: 802 LIATGNIDTNAVIFD-RPSGQVLATLTGHSKKVTSVKFVGQGESIITGSADKTVRLWQGS 978
Query: 277 AVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVD 336
+ + L +H A V+A+ + + G C +L + +++
Sbjct: 979 DDGHYNCKQIL---KDHSAEVEAVTVHATNNYFVTASLDGSWCFYELSSGTCLTQVSDSS 1149
Query: 337 TGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGST 396
G A + L+ G T + + +W S +A+ GH V ++ S +G
Sbjct: 1150EGYTSAAF----HPDGLILGTGTTDSLVKIWDVKSQANVAKFDGHVGHVTAISFSENGYY 1317
Query: 397 VASAA 401
+A+AA
Sbjct: 1318LATAA 1332
Score = 42.4 bits (98), Expect = 4e-04
Identities = 26/78 (33%), Positives = 37/78 (47%), Gaps = 2/78 (2%)
Frame = +1
Query: 150 SSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKG--GHRAR 207
SSG+ V + TS + PDG L G T+S V++WD + + G GH
Sbjct: 1111 SSGTCLTQVSDSSEGYTSAAFHPDGLILGTGTTDSLVKIWDVKSQANVAKFDGHVGHVTA 1290
Query: 208 VGSLAWNGHVLTTGGMDG 225
+ S + NG+ L T DG
Sbjct: 1291 I-SFSENGYYLATAAHDG 1341
Score = 40.0 bits (92), Expect = 0.002
Identities = 42/182 (23%), Positives = 78/182 (42%), Gaps = 7/182 (3%)
Frame = +1
Query: 151 SGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA--ANKQLRTLKGGHRARV 208
SG T+ VTSV + G + G + V+LW + + + + H A V
Sbjct: 853 SGQVLATLTGHSKKVTSVKFVGQGESIITGSADKTVRLWQGSDDGHYNCKQILKDHSAEV 1032
Query: 209 GSLAWN--GHVLTTGGMDGKIVNNDV---RLRSQIINTYRGHRREVCGLKWSLDGKQLAS 263
++ + + T +DG ++ +Q+ ++ G+ + DG L +
Sbjct: 1033 EAVTVHATNNYFVTASLDGSWCFYELSSGTCLTQVSDSSEGY----TSAAFHPDGLILGT 1200
Query: 264 GGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKL 323
G D++V IWD+ + ++ + +FD H V A+++ G LA+ G VKL
Sbjct: 1201 GTTDSLVKIWDVKSQAN------VAKFDGHVGHVTAISFSE-NGYYLATAAHDG---VKL 1350
Query: 324 WN 325
W+
Sbjct: 1351 WD 1356
>TC78914 similar to GP|13937195|gb|AAK50091.1 At1g78070/F28K19_28 {Arabidopsis
thaliana}, partial (76%)
Length = 1732
Score = 48.5 bits (114), Expect = 5e-06
Identities = 38/112 (33%), Positives = 53/112 (46%), Gaps = 2/112 (1%)
Frame = +1
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW--NGHVLTTGG 222
V + +PDG+ LAV ++ + D K LKG H S AW NG +L TG
Sbjct: 931 VNNASVSPDGKLLAVLGDSTEGLIADANTGKVTGNLKG-HLDYSFSSAWHPNGQILATGN 1107
Query: 223 MDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWD 274
D D+R S+ I +G + LK++ DGK LA + VHI+D
Sbjct: 1108 QDTTCRLWDIRNLSESIAVLKGRMGAIRSLKFTSDGKFLAMAEPADFVHIFD 1263
Score = 30.0 bits (66), Expect = 1.8
Identities = 29/132 (21%), Positives = 51/132 (37%), Gaps = 3/132 (2%)
Frame = +1
Query: 153 SEFVTVDEEEGPVTSVCWAPDGRHLAVGLTN-SHVQLWDTAANKQLRTLKGGHRARVGSL 211
S+ T D V P G + N S V+++D L K S+
Sbjct: 769 SKITTDDSAITNAVDVFRNPRGSLKVIAANNDSQVRVFDAENFASLGCFKYDWSVNNASV 948
Query: 212 AWNGHVLTTGG--MDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNV 269
+ +G +L G +G I + + ++ +GH W +G+ LA+G D
Sbjct: 949 SPDGKLLAVLGDSTEGLIADANT---GKVTGNLKGHLDYSFSSAWHPNGQILATGNQDTT 1119
Query: 270 VHIWDMSAVSSN 281
+WD+ +S +
Sbjct: 1120CRLWDIRNLSES 1155
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,309,057
Number of Sequences: 36976
Number of extensions: 259050
Number of successful extensions: 2704
Number of sequences better than 10.0: 146
Number of HSP's better than 10.0 without gapping: 2172
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2586
length of query: 436
length of database: 9,014,727
effective HSP length: 99
effective length of query: 337
effective length of database: 5,354,103
effective search space: 1804332711
effective search space used: 1804332711
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146343.9


