

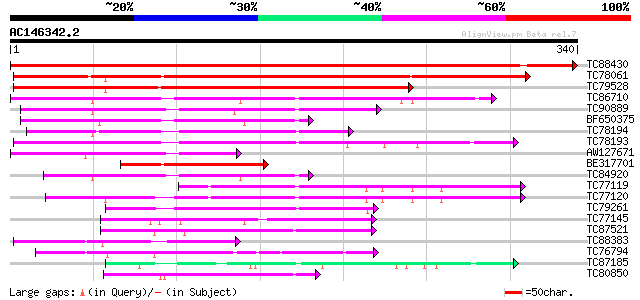
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146342.2 + phase: 0
(340 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protei... 641 0.0
TC78061 weakly similar to PIR|T10824|T10824 auxin-induced protei... 271 3e-73
TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone ox... 226 7e-60
TC86710 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 131 3e-31
TC90889 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 114 5e-26
BF650375 similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabido... 99 2e-21
TC78194 similar to PIR|T05166|T05166 quinone reductase homolog F... 96 2e-20
TC78193 similar to PIR|T05166|T05166 quinone reductase homolog F... 95 4e-20
AW127671 weakly similar to GP|15724256|gb AT4g13010/F25G13_100 {... 91 6e-19
BE317701 similar to GP|14532287|gb| quinone oxidoreductase-like ... 87 1e-17
TC84920 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 85 3e-17
TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 79 3e-15
TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 78 6e-15
TC79261 similar to PIR|T49047|T49047 quinone reductase-like prot... 75 3e-14
TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehyd... 57 1e-08
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 57 1e-08
TC88383 similar to GP|21592515|gb|AAM64465.1 nuclear receptor bi... 56 2e-08
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 52 3e-07
TC87185 weakly similar to GP|20161259|dbj|BAB90185. putative all... 52 3e-07
TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehyd... 49 2e-06
>TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protein (clone
MII-3) - mung bean, partial (41%)
Length = 1450
Score = 641 bits (1653), Expect = 0.0
Identities = 325/341 (95%), Positives = 328/341 (95%), Gaps = 1/341 (0%)
Frame = +2
Query: 1 MQKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAV 60
MQKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAV
Sbjct: 29 MQKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAV 208
Query: 61 PGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK 120
PGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK
Sbjct: 209 PGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK 388
Query: 121 RLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVS 180
RLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVS
Sbjct: 389 RLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVS 568
Query: 181 SCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDIT 240
SCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDIT
Sbjct: 569 SCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDIT 748
Query: 241 WPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAW 300
WPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAW
Sbjct: 749 WPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAW 928
Query: 301 GKVVVTCFPMAPKDRHSTILSEIDTNIPQVNGF-AKDLCLK 340
GKVVVT P +TILSEIDTN Q+NG AK+LCLK
Sbjct: 929 GKVVVT----IPHHHSTTILSEIDTNTLQLNGLAAKNLCLK 1039
>TC78061 weakly similar to PIR|T10824|T10824 auxin-induced protein (clone
MII-3) - mung bean, complete
Length = 1315
Score = 271 bits (692), Expect = 3e-73
Identities = 144/315 (45%), Positives = 207/315 (65%), Gaps = 5/315 (1%)
Frame = +3
Query: 3 KAWFYEEYGP-KEVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD---F 57
KAWFY E+G ++LKL ++ IP ++Q+L++V AA+LNP+D KR M +F
Sbjct: 117 KAWFYSEHGKASDILKLHPNWSIPQLKDDQVLIKVVAASLNPVDYKR-MHALFKETDPHL 293
Query: 58 PAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVAR 117
P VPG D+AG+VI G V KF +GDE+YG+I + + K LGTL+++ + EE L+A
Sbjct: 294 PIVPGFDVAGIVIKVGSEVVKFKVGDEIYGDINE-EGLSNLKILGTLSEYTIAEERLLAH 470
Query: 118 KPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASY 177
KPK LSF EAAS+PLA++TA EG + G+++ V+GGAGGVG+ +QLAK ++GAS
Sbjct: 471 KPKNLSFIEAASIPLAMETAYEGLERAQLSAGKSILVLGGAGGVGSFAIQLAKHVYGASK 650
Query: 178 VVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIV 237
+ ++ ST K++F+++ G D +DYTK +ED+ EKFD +YD VG+ ++ K+ G +V
Sbjct: 651 IAATSSTGKIEFLRKLGVDLPIDYTKENFEDLPEKFDVVYDGVGEVDRAMKAIKEGGKVV 830
Query: 238 DITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETG 297
I P A++ LT G ILEKLRPY E G+LK ++D K F VI+A+ Y+ET
Sbjct: 831 TIV-PPGFPPAIFFVLTSKGSILEKLRPYFESGQLKPILDSKTPVPFSEVIEAYSYLETS 1007
Query: 298 RAWGKVVVTCFPMAP 312
RA GKVV+ P P
Sbjct: 1008RATGKVVIYPIP*NP 1052
>TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone
oxidoreductase {Fragaria x ananassa}, partial (72%)
Length = 834
Score = 226 bits (577), Expect = 7e-60
Identities = 121/244 (49%), Positives = 166/244 (67%), Gaps = 4/244 (1%)
Frame = +2
Query: 3 KAWFYEEYGPK-EVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FP 58
KAW Y EYG +VLK + PIP ++Q+L++V AA+LNPID KR SD P
Sbjct: 92 KAWTYSEYGHSVDVLKFDPNVPIPQVKDDQVLIKVAAASLNPIDYKRMEGAFKASDSPLP 271
Query: 59 AVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARK 118
PG D++GVV+ G VKKF +GDEVYG+I + ++E PK +G+LA++ EE L+A K
Sbjct: 272 TAPGYDVSGVVVKVGSEVKKFKVGDEVYGDI-NVKALEYPKVIGSLAEYTAAEEILLAHK 448
Query: 119 PKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
PK LSF EAASLPL ++TA EG + F G+++ V+GGAGGVGT V+QLAK ++GAS V
Sbjct: 449 PKNLSFAEAASLPLTIETAYEGLERTGFSAGKSILVLGGAGGVGTHVIQLAKHVYGASKV 628
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVD 238
++ ST KL+ + GAD +DYTK +ED+ EKFD ++DTVG+ +K+F K+ G +V
Sbjct: 629 AATSSTKKLELLSNLGADLPIDYTKENFEDLPEKFDVVFDTVGETEKAFKALKEGGKVVT 808
Query: 239 ITWP 242
I P
Sbjct: 809 IVPP 820
>TC86710 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (98%)
Length = 1176
Score = 131 bits (330), Expect = 3e-31
Identities = 105/322 (32%), Positives = 155/322 (47%), Gaps = 30/322 (9%)
Frame = +3
Query: 1 MQKAWFYEEYGPK-EVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIF-PS 55
+ +A Y YG LK + P+P P N++L+++ A ++NPID K + +R IF P
Sbjct: 54 LMQALQYSSYGGGVSGLKHAEVPVPIPKTNEVLLKLEATSINPIDWKIQSGALRAIFLPR 233
Query: 56 DFPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLV 115
FP +P D+AG V+ G VK F GD+V + + G LA+F V E+L
Sbjct: 234 KFPHIPCTDVAGEVVEVGPQVKDFKSGDKVIAKLTH-------QYGGGLAEFAVASESLT 392
Query: 116 ARKPKRLSFEEAASLPLAVQTA------IEGFKTGDFKKGETMFVVGGAGGVGTLVLQLA 169
A +P +S EAA LP+A TA I G K + + + V +GGVG+ +QLA
Sbjct: 393 ATRPPEVSAAEAAGLPIAGLTARDALTEIGGIKLDGTGEQKNVLVTAASGGVGSFAVQLA 572
Query: 170 KLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVV 229
KL G ++V ++C + FVK GAD+V+DY + ++ YD V C
Sbjct: 573 KL--GNNHVTATCGARNIDFVKSLGADEVLDYKTPEGTSLKSPSGKKYDAVIHCTTGIPW 746
Query: 230 AKKD------GAIVDIT-------------WPASHERAVYSSLTVCGEILEKLRPYLERG 270
+ D G +VD+T S +R V +TV E LE L ++ G
Sbjct: 747 STFDPNLSEKGVVVDLTPGPSSMLTFALKKTTFSKKRLVPFVVTVKREGLEHLAQLVKDG 926
Query: 271 ELKAVIDPKGEYDFENVIDAFG 292
+LK +ID K + DA G
Sbjct: 927 KLKTIIDSK--FPLSKAEDALG 986
>TC90889 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (69%)
Length = 779
Score = 114 bits (285), Expect = 5e-26
Identities = 73/227 (32%), Positives = 117/227 (51%), Gaps = 10/227 (4%)
Frame = +3
Query: 7 YEEY-GPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPAVPG 62
Y+ Y G LK + IP+P N++L++V A+++NP+D K + +R + P FP P
Sbjct: 75 YDSYDGGSSGLKHVEVHIPTPKANEVLIKVEASSINPVDWKIQDGLLRYLLPRKFPFTPC 254
Query: 63 CDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRL 122
D+AG V+ G VK F +GD+V + + + G LA+F V E+L +P +
Sbjct: 255 TDVAGEVVDFGSQVKDFKVGDKVIAKLNNQDG-------GGLAEFAVASESLTTLRPSEV 413
Query: 123 SFEEAASLPLA------VQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGAS 176
S E A LP+A T + G K + + + V +GGVG +QLAKL G +
Sbjct: 414 SAAEGAGLPVAGLAAHDAITKMAGIKLDRTGEPKNILVTAASGGVGVYAVQLAKL--GNN 587
Query: 177 YVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDC 223
+V ++C ++ +K GAD+V+DY + ++ YD V C
Sbjct: 588 HVTATCGARNIELIKSLGADEVLDYKTPEGASLKSPSGRKYDAVIHC 728
>BF650375 similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabidopsis
thaliana}, partial (55%)
Length = 564
Score = 99.0 bits (245), Expect = 2e-21
Identities = 66/187 (35%), Positives = 100/187 (53%), Gaps = 11/187 (5%)
Frame = +1
Query: 7 YEEYGP-KEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI----FPSDFPAVP 61
Y YG LK + PIP+P N++L+++ A ++NP+D K + + FP FP +P
Sbjct: 25 YNSYGGGASGLKHVEVPIPTPKTNEVLIKLEAISINPLDWKIQQGVLRAMFFPRKFPHIP 204
Query: 62 GCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKR 121
D+AG V+ G VK F +GD+V + + + G LA+F V E+L A +P
Sbjct: 205 CTDVAGEVVEVGPRVKDFKVGDKVIAKLTN-------EYGGGLAEFAVASESLTAARPSE 363
Query: 122 LSFEEAASLPLAVQTAIE------GFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGA 175
+S EAA LP+A TA + G K + + + V +GGVG V+QLAKL G
Sbjct: 364 VSAAEAAGLPIAGITAHDALTKNGGIKLDGTGEQKNILVTAASGGVGAYVVQLAKL--GN 537
Query: 176 SYVVSSC 182
++V +C
Sbjct: 538 NHVTXTC 558
>TC78194 similar to PIR|T05166|T05166 quinone reductase homolog F18E5.200 -
Arabidopsis thaliana, partial (60%)
Length = 709
Score = 95.9 bits (237), Expect = 2e-20
Identities = 66/199 (33%), Positives = 99/199 (49%), Gaps = 3/199 (1%)
Frame = +1
Query: 11 GPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR--MRPIFPSDFPAVPGCDMAGV 68
G EVL+L + P P +N++L++V+A ALN D+ +R PI P PG + +G
Sbjct: 148 GGPEVLQLQEVQDPQPKDNEVLIRVHATALNSADTVQRKGFYPI-PQGASPYPGLECSGT 324
Query: 69 VIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAA 128
++ G+NV K+ IGD+V + G A+ + V E V P +S ++AA
Sbjct: 325 IVCVGINVSKWKIGDQVCALLAG----------GGYAEKVAVPEGQVLPVPPGISLKDAA 474
Query: 129 SLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKL 187
S T F T KGET+ V GG+ G+GT + +AK G+ V++ S KL
Sbjct: 475 SFTEVACTVWSTIFMTSRLSKGETLLVHGGSSGIGTFAIXIAKYQ-GSKVFVTAGSEEKL 651
Query: 188 KFVKQFGADKVVDYTKTKY 206
F K GAD +Y +
Sbjct: 652 AFCKSIGADVGXNYXTXDF 708
>TC78193 similar to PIR|T05166|T05166 quinone reductase homolog F18E5.200 -
Arabidopsis thaliana, partial (95%)
Length = 1379
Score = 94.7 bits (234), Expect = 4e-20
Identities = 84/335 (25%), Positives = 148/335 (44%), Gaps = 32/335 (9%)
Frame = +2
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPA-VP 61
KA + G EVL+L + P ++++L++V+A ALN D+ +R P + P
Sbjct: 20 KAIVITKPGEPEVLQLQEVQDPQIKDDEVLIKVHATALNRADTLQRKGAYPPPQGASPYP 199
Query: 62 GCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKR 121
G + +G++ G NV K+ IGD+V + G A+ + V E V P
Sbjct: 200 GLECSGIIEFVGKNVSKWKIGDQVCALLAG----------GGYAEKVAVAEGQVLPVPPG 349
Query: 122 LSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVS 180
+S ++AAS P T F T KGET+ + GG+ G+GT +Q+AK + G+ V+
Sbjct: 350 ISLKDAASFPEVACTVWSTIFMTSRLSKGETLLIHGGSSGIGTFAIQIAKYL-GSRVFVT 526
Query: 181 SCSTPKLKFVKQFGADKVVDY------TKTKYEDIEEKFDFIYDTVGDC----------- 223
+ S KL F K GAD ++Y + K E + D I D +G
Sbjct: 527 AGSEEKLAFCKSIGADVGINYKTEDFVARVKEETGGQGVDVILDCMGASYYQRNLASLNF 706
Query: 224 -KKSFVVAKKDGAIVDITWPA------------SHERAVYSSLTVCGEILEKLRPYLERG 270
+ F++ + G ++ A R+ + + E+ + + P + G
Sbjct: 707 DGRLFIIGFQGGVSTELDLRALFGKRLTVQAAGLRSRSPENKAVIVAEVEKNVWPAIAEG 886
Query: 271 ELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
++K V+ + +A +E+ + GK+++
Sbjct: 887 KVKPVV--YKSFPLSEAAEAHRLMESSQHIGKILL 985
>AW127671 weakly similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabidopsis
thaliana}, partial (48%)
Length = 493
Score = 90.9 bits (224), Expect = 6e-19
Identities = 54/143 (37%), Positives = 82/143 (56%), Gaps = 4/143 (2%)
Frame = +2
Query: 1 MQKAWFYEEYGPKEV-LKLGDFPIPSPLENQLLVQVYAAALNPID---SKRRMRPIFPSD 56
+ +A Y YG LK + PIPSP ++++L+++ AA++NP D KR + P P
Sbjct: 20 LMRAVQYNAYGGGSTGLKHVEVPIPSPSKDEVLIKLEAASINPFDWKVQKRMLWPFLPPK 199
Query: 57 FPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVA 116
FP +P D+AG V+ G VKKF GD+V G + F+ G LA+F VV+E++ A
Sbjct: 200 FPYIPCTDVAGEVMMVGKGVKKFKTGDKVVGLVSPFSG-------GGLAEFAVVKESVTA 358
Query: 117 RKPKRLSFEEAASLPLAVQTAIE 139
P+ +S E LP+A TA++
Sbjct: 359 SIPQEISASECVGLPVAGLTALQ 427
>BE317701 similar to GP|14532287|gb| quinone oxidoreductase-like protein
{Helianthus annuus}, partial (28%)
Length = 269
Score = 86.7 bits (213), Expect = 1e-17
Identities = 42/89 (47%), Positives = 61/89 (68%)
Frame = +3
Query: 67 GVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEE 126
GVV+ G VK F +GDEVYG++ + ++E P Q G+LA++ VEE L+A KP L F +
Sbjct: 3 GVVVKVGSEVKDFKVGDEVYGDVNE-KALEGPNQFGSLAEYTAVEEKLLALKPTNLDFAQ 179
Query: 127 AASLPLAVQTAIEGFKTGDFKKGETMFVV 155
AASLPLA++TA EG + F G+++ V+
Sbjct: 180 AASLPLAIETAYEGLERTGFSSGKSILVL 266
>TC84920 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (54%)
Length = 563
Score = 85.1 bits (209), Expect = 3e-17
Identities = 60/172 (34%), Positives = 88/172 (50%), Gaps = 10/172 (5%)
Frame = +2
Query: 21 FPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIF-PSDFPAVPGCDMAGVVIGKGVNV 76
F P N++L+++ A +NP D K + +R IF P FP P D+AG V+ G V
Sbjct: 53 FLFQPPKTNEVLIKLEAIRINPADWKIQTGVLRAIFFPRKFPHTPCTDVAGEVVEIGEQV 232
Query: 77 KKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQT 136
K F +GD+V + + G LA+F V E+L A +P +S EAA LP+A T
Sbjct: 233 KDFKVGDKVIAKLNNLYG-------GGLAEFAVARESLTAARPSEVSAAEAAGLPIAGLT 391
Query: 137 A------IEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSC 182
A I G K + + + V +GGV +QLAKL G ++V ++C
Sbjct: 392 ARDALTEIGGVKLDGTGEPKNVLVTAASGGVXVYAVQLAKL--GNNHVTATC 541
>TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2277
Score = 79.0 bits (193), Expect = 3e-15
Identities = 69/242 (28%), Positives = 105/242 (42%), Gaps = 34/242 (14%)
Frame = +1
Query: 102 GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGV 161
G A+F ++ P R E A L + +I K G + G+ + V AGG
Sbjct: 1249 GGYAEFTMIPSKYALPVP-RPDPEGVAMLTSGLTASIALEKAGQMESGKVVLVTAAAGGT 1425
Query: 162 GTLVLQLAKLMFGASYVVSSCST-PKLKFVKQFGADKVVDYTKTKYEDIEEK-----FDF 215
G +QLAKL + VV++C K K +K+ G D+V+DY + + K D
Sbjct: 1426 GQFAVQLAKL--AGNTVVATCGGGTKAKLLKELGVDRVIDYNSEDIKTVLRKEFPKGIDI 1599
Query: 216 IYDTVGD-----CKKSFVVAKKDGAIVDIT------------WPASHERAVYSSLTVCG- 257
IY++VG C + V + I I+ +P E+ + S TV G
Sbjct: 1600 IYESVGGDMLKLCLDALAVHGRLIVIGMISQYQGEHGWTPSKYPGLLEKLLAKSQTVAGF 1779
Query: 258 ----------EILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVVTC 307
E L++L +G+LK +DPK +V DA Y+ +G++ GKVVV
Sbjct: 1780 FLVQYSHFYQEHLDRLFDLYSKGKLKVAVDPKKFIGLHSVADAVEYLHSGKSVGKVVVCV 1959
Query: 308 FP 309
P
Sbjct: 1960 DP 1965
>TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2203
Score = 77.8 bits (190), Expect = 6e-15
Identities = 83/330 (25%), Positives = 133/330 (40%), Gaps = 42/330 (12%)
Frame = +3
Query: 22 PIPSPLE-NQLLVQVYAAALNPIDSKRRMRPIFPSD-------FPAVPGCDMAGVVIGKG 73
P+ P++ N +LV++ A +N D F + P G + G++ G
Sbjct: 1044 PLRLPIKPNHVLVKIIYAGVNASDVNFSSGRYFGGNNKETAARLPFDAGFEAVGIIAAVG 1223
Query: 74 VNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLA 133
+V +G G A+F ++ P R E A L
Sbjct: 1224 DSVTDLKVGMPCAF-----------MTFGGYAEFTMIPSKYALPVP-RPDPEAVAMLTSG 1367
Query: 134 VQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCST-PKLKFVKQ 192
+ +I K G + G+ + V AGG G +QLAKL + VV++C K K +K+
Sbjct: 1368 LTASIALEKAGQMESGKVVLVTAAAGGTGQFAVQLAKL--AGNTVVATCGGGAKAKLLKE 1541
Query: 193 FGADKVVDYTKTKYEDIEEK-----FDFIYDTVGD-----CKKSFVVAKKDGAIVDIT-- 240
G D+V+DY + + +K D IY++VG C + V + I I+
Sbjct: 1542 LGVDRVIDYHSEDIKTVLKKEFPKGIDIIYESVGGDMLKLCLDALAVHGRLIVIGMISQY 1721
Query: 241 ----------WPASHERAVYSSLTVCG-----------EILEKLRPYLERGELKAVIDPK 279
+P E+ + S V G E L++L +G+LK +DPK
Sbjct: 1722 QGEKGWTPSKYPGLCEKLLSKSQAVAGFFLVQYSHMWQEHLDRLFDLYSQGKLKVAVDPK 1901
Query: 280 GEYDFENVIDAFGYIETGRAWGKVVVTCFP 309
+V DA Y+ +G++ GKVVV P
Sbjct: 1902 KFIGLHSVADAVEYLHSGKSAGKVVVCVDP 1991
>TC79261 similar to PIR|T49047|T49047 quinone reductase-like protein -
Arabidopsis thaliana, partial (95%)
Length = 1239
Score = 75.5 bits (184), Expect = 3e-14
Identities = 52/174 (29%), Positives = 80/174 (45%), Gaps = 10/174 (5%)
Frame = +3
Query: 58 PAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVAR 117
P +PG D +G+V G NV F +GD V LG+ AQ++VV++ + R
Sbjct: 228 PFIPGSDFSGIVDSVGSNVTNFRVGDPVCSF----------AALGSYAQYLVVDQTQLFR 377
Query: 118 KPKRLSFEEAASLPLAVQTAIEGF-KTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGAS 176
P+ A +L +A T+ G K G+ + V+G AGGVG +Q+ K GA
Sbjct: 378 VPEGCDLVAAGALAVAFGTSHVGLVHRAQLKSGQVLLVLGAAGGVGLAAVQIGKAC-GAI 554
Query: 177 YVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKF---------DFIYDTVG 221
+ + K++ +K G D VVD + ++F D +YD VG
Sbjct: 555 VIAVARGAEKVQLLKSMGVDHVVDLGNENVTESVKEFLKVKRLKGIDVLYDPVG 716
>TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) (CAD). [Alfalfa] {Medicago sativa},
complete
Length = 1384
Score = 57.0 bits (136), Expect = 1e-08
Identities = 54/197 (27%), Positives = 87/197 (43%), Gaps = 31/197 (15%)
Frame = +1
Query: 55 SDFPAVPGCDMAGVVIGKGVNVKKFDIGD---------------------EVYGN--IQD 91
S++P VPG ++ G V+ G NV +F +G+ E Y N I
Sbjct: 235 SNYPMVPGHEVVGEVLEVGSNVTRFKVGEIVGVGLLVGCCKSCRACDSEIEQYCNKKIWS 414
Query: 92 FNSMEKPKQL--GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIE-----GFKTG 144
+N + ++ G A+ VVE+ V + P+ L+ E+ A L A T G KT
Sbjct: 415 YNDVYTDGKITQGGFAESTVVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLSHFGLKTP 594
Query: 145 DFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTK 203
+ G + G GGVG + +++AK V+SS K + ++ GAD +V
Sbjct: 595 GLRGG-----ILGLGGVGHMGVKVAKAFGHHVTVISSSDKKKKEALEDLGADSYLVSSDT 759
Query: 204 TKYEDIEEKFDFIYDTV 220
++ + D+I DTV
Sbjct: 760 VGMQEAADSLDYIIDTV 810
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 56.6 bits (135), Expect = 1e-08
Identities = 56/193 (29%), Positives = 82/193 (42%), Gaps = 27/193 (13%)
Frame = +2
Query: 55 SDFPAVPGCDMAGVVIGKGVNVKKFDIGDEV-----------YGNIQDFNSMEKPKQLGT 103
S +P VPG ++AG+V G V+KF IGD+V N ++ PKQ T
Sbjct: 257 STYPLVPGHEIAGIVTEVGSKVEKFKIGDKVGVGCLVDSCRACQNCEENLENYCPKQTNT 436
Query: 104 --------------LAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKG 149
+ +V +E+ + P L E AA L A T + K
Sbjct: 437 YSAKYSDGSITYGGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGITVYSPLRYFGLDKP 616
Query: 150 ETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLK-FVKQFGADK-VVDYTKTKYE 207
+ G GG+G L ++ AK FGA+ V S S K K ++ GAD ++ + + K +
Sbjct: 617 GMNIGIVGLGGLGHLGVKFAK-AFGANVTVISTSPNKEKEAIENLGADSFLISHDQDKMQ 793
Query: 208 DIEEKFDFIYDTV 220
D I DTV
Sbjct: 794 AAMGTLDGIIDTV 832
>TC88383 similar to GP|21592515|gb|AAM64465.1 nuclear receptor binding
factor-like protein {Arabidopsis thaliana}, partial
(90%)
Length = 1313
Score = 55.8 bits (133), Expect = 2e-08
Identities = 46/139 (33%), Positives = 67/139 (48%), Gaps = 3/139 (2%)
Frame = +3
Query: 3 KAWFYEEYG-PKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFP--SDFPA 59
KA YE +G P V KL D P EN L V++ AA +NP D R++ ++P + PA
Sbjct: 126 KAIIYESHGQPDAVTKLVDIPATEVKENDLCVKMLAAPINPSDI-NRIQGVYPVRPEPPA 302
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
V G + G V G V F GD V + P GT +IV ++N+ +
Sbjct: 303 VGGYEGVGEVHSVGSAVTCFSPGDWV---------IPSPPSFGTWQTYIVKDQNVWHKVN 455
Query: 120 KRLSFEEAASLPLAVQTAI 138
K + E AA++ + TA+
Sbjct: 456 KGVPMEYAATITVNPLTAL 512
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 52.4 bits (124), Expect = 3e-07
Identities = 62/230 (26%), Positives = 98/230 (41%), Gaps = 24/230 (10%)
Frame = +2
Query: 16 LKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDF----PAVPGCDMAGVVIG 71
LK+ F +PS + + +++ A + D ++ + +DF P V G + AG++
Sbjct: 293 LKIQPFNLPSLGPHDVRIKMKAVGICGSDV-HYLKTLRCADFIVKEPMVIGHECAGIIEE 469
Query: 72 KGVNVKKFDIGDEV------------------YGNIQDFNSMEKPKQLGTLAQFIVVEEN 113
G VK GD V Y D P G+LA IV +
Sbjct: 470 VGSQVKTLVPGDRVAIEPGISCWRCDHCKLGRYNLCPDMKFFATPPVHGSLANQIVHPAD 649
Query: 114 LVARKPKRLSFEEAASL-PLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLM 172
L + P+ +S EE A PL+V + + + ET ++ GAG +G LV L+
Sbjct: 650 LCFKLPENVSLEEGAMCEPLSV--GVHACRRANIGP-ETNVLIMGAGPIG-LVTMLSARA 817
Query: 173 FGAS-YVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVG 221
FGA VV +L K GAD +V T +D+ E+ I++ +G
Sbjct: 818 FGAPRIVVVDVDDHRLSVAKSLGADDIVK-VSTNIQDVAEEVKQIHNVLG 964
>TC87185 weakly similar to GP|20161259|dbj|BAB90185. putative allyl alcohol
dehydrogenase {Oryza sativa (japonica cultivar-group)},
partial (43%)
Length = 1243
Score = 52.0 bits (123), Expect = 3e-07
Identities = 80/292 (27%), Positives = 109/292 (36%), Gaps = 44/292 (15%)
Frame = +2
Query: 58 PAVPGCDMAGVVIGKGVNV--KKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLV 115
P PG + +VIG+ V KF+ D V G + T A++ VV+E +
Sbjct: 266 PLTPGQAIDALVIGRVVTSGNAKFEKDDLVMG-------------VFTWAEYSVVKEQSI 406
Query: 116 ARKPKRLSFEEAASLPLAVQTAIEGFKT----GDF------KKGETMFVVGGAGGVGTLV 165
+K + F PL I GF G F +KGET+FV +G VG +V
Sbjct: 407 IKKLESFEF------PLTYHLGILGFSGLSAYGGFFEICKPRKGETVFVSAASGSVGNIV 568
Query: 166 LQLAKLMFGASYVVSSCSTPK--------LKFVKQFGADKVVDYTKTKYEDIEEKFDFIY 217
Q AKL+ YVV + K L F F + D T + D +
Sbjct: 569 GQYAKLL--GCYVVGCAGSQKKVTLLKEELGFDDAFNYKEETDLNSTLKRYFPDGIDIYF 742
Query: 218 DTVGDCKKSFVVA--KKDGAI---------VDITWPASHER--AVYSSLT---------- 254
D VG VA K G + DI AS VY +T
Sbjct: 743 DNVGGEMLEAAVANMKAFGRVSVCGVISEYTDIGKRASPNMMDVVYKRITIRGFLAADYM 922
Query: 255 -VCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
V G+ K YL G+L+ + D E++ AF + G GK VV
Sbjct: 923 NVFGDFSAKTLDYLRNGQLRVIED--RSLGVESIPSAFVGLFNGDNVGKKVV 1072
>TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Alfalfa], partial (61%)
Length = 710
Score = 49.3 bits (116), Expect = 2e-06
Identities = 44/154 (28%), Positives = 63/154 (40%), Gaps = 24/154 (15%)
Frame = +1
Query: 57 FPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNI------------QD-----------FN 93
+P VPG ++ GVV G+NVKKF +GD V + QD +N
Sbjct: 247 YPVVPGHEIVGVVTKVGINVKKFRVGDNVGVGVIVESCQTCENCNQDLEQYCPKLVFTYN 426
Query: 94 SMEK-PKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETM 152
S K + G + F+VV + V + P L + A L A T K +
Sbjct: 427 SPYKGTRTHGGYSDFVVVHQRYVVQFPDNLPLDAGAPLLCAGITVYSPMKYYGMTEPGKH 606
Query: 153 FVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPK 186
V G GG+G + ++ K FG V + S K
Sbjct: 607 LGVAGLGGLGHVAIKFGK-AFGLKVTVITTSPNK 705
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.139 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,163,818
Number of Sequences: 36976
Number of extensions: 111618
Number of successful extensions: 515
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 485
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 490
length of query: 340
length of database: 9,014,727
effective HSP length: 97
effective length of query: 243
effective length of database: 5,428,055
effective search space: 1319017365
effective search space used: 1319017365
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146342.2


