

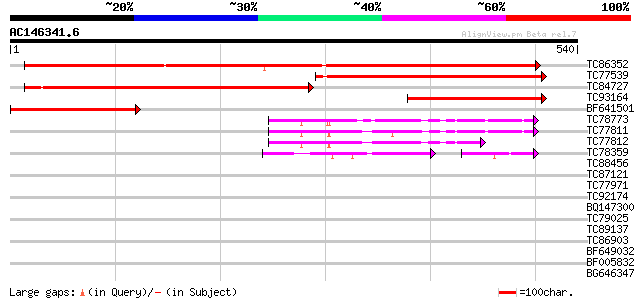
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.6 + phase: 0
(540 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86352 similar to PIR|H86192|H86192 hypothetical protein [impor... 476 e-135
TC77539 similar to GP|16118856|gb|AAL14629.1 growth-on protein G... 351 4e-97
TC84727 similar to GP|16118856|gb|AAL14629.1 growth-on protein G... 346 1e-95
TC93164 similar to GP|16118854|gb|AAL14628.1 growth-on protein G... 264 6e-71
BF641501 similar to GP|16118854|gb| growth-on protein GRO11 {Eup... 256 2e-68
TC78773 similar to GP|17473842|gb|AAL38345.1 unknown protein {Ar... 100 1e-21
TC77811 similar to GP|17473842|gb|AAL38345.1 unknown protein {Ar... 86 5e-17
TC77812 similar to GP|17473842|gb|AAL38345.1 unknown protein {Ar... 72 7e-13
TC78359 similar to GP|16648809|gb|AAL25595.1 AT4g33410/F17M5_170... 70 2e-12
TC88456 similar to GP|15487292|dbj|BAB64531. vacuolar sorting re... 41 0.001
TC87121 weakly similar to PIR|T06794|T06794 vacuolar sorting rec... 40 0.003
TC77971 similar to PIR|T06794|T06794 vacuolar sorting receptor p... 38 0.011
TC92174 homologue to PIR|T06794|T06794 vacuolar sorting receptor... 35 0.095
BQ147300 weakly similar to PIR|T01080|T01 hypothetical protein T... 32 0.81
TC79025 similar to PIR|T49099|T49099 dihydrolipoamide S-acetyltr... 31 1.1
TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycop... 31 1.4
TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylas... 31 1.4
BF649032 weakly similar to PIR|T45951|T459 hypothetical protein ... 30 2.3
BF005832 similar to PIR|T05571|T055 hypothetical protein F22K18.... 29 5.2
BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.22... 28 6.8
>TC86352 similar to PIR|H86192|H86192 hypothetical protein [imported] -
Arabidopsis thaliana, partial (37%)
Length = 2150
Score = 476 bits (1226), Expect = e-135
Identities = 243/493 (49%), Positives = 325/493 (65%), Gaps = 2/493 (0%)
Frame = +3
Query: 15 VFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLE 74
+ L + + + D+ DD AP C N F LVKV W DG E G+ ARFG +L
Sbjct: 234 LLLLATTAFASNDVKRDDDRAPKSKTCNNPFQLVKVKNWADGEEGVIQSGMTARFGSSLP 413
Query: 75 SKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIIN 134
K +++ TRV+ ++P DCCS +KL + + L RG C F KA IA GA+ +LIIN
Sbjct: 414 EKAENSVRTRVLFSNPTDCCSSSTSKLVDSVALCIRGGCDFQLKATIAQSGGATGVLIIN 593
Query: 135 YRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAE 194
+L +MVC ++ T+ +I IP VM+ + AG L + + V + LY+P RPLVD +
Sbjct: 594 NEEDLVEMVCTDS-TEANITIPVVMITKSAGEALNTSLTSGKRVDVLLYAPPRPLVDFSV 770
Query: 195 VFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGS--RGYVEISTTAA 252
FLWL++VGTI+CAS WS T E + E+ L + VA + G + + I++ AA
Sbjct: 771 AFLWLVSVGTIVCASLWSDLTTPEKSGERYSELYPKESQNVAAARGGSDKQVLNINSKAA 950
Query: 253 ILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYV 312
++FV+ AS FLV+L+ MS WF+ +L+VLFCI GIEG+ C+T+L R +Q Q V
Sbjct: 951 VVFVITASTFLVLLFFFMSSWFMWLLIVLFCIAGIEGMHNCITSLT--LRKWQNIGQKTV 1124
Query: 313 KIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLK 372
+P FG + +LAV FC+ FAV WA R SY+WIGQD LGI L+ITVLQ+ ++PN+K
Sbjct: 1125NVPLFGEISIFSLAVFLFCLAFAVFWAATRHESYSWIGQDTLGICLMITVLQLAQLPNIK 1304
Query: 373 VGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYS 432
V TVLLSCAF YDI WVF+S FHESVMI VARGDK+G + IPMLL+ PRLFD WGGY
Sbjct: 1305VATVLLSCAFAYDIFWVFISPLIFHESVMIAVARGDKAGGEAIPMLLRFPRLFDTWGGYD 1484
Query: 433 IIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQP 492
+IGFGDII PGL+V+F+ R+D KK + GYF+W + YG+GL+ TY+ L LMDG+GQP
Sbjct: 1485MIGFGDIIFPGLLVSFAHRFDKDNKKGVLNGYFLWLVIGYGIGLVFTYLGLYLMDGNGQP 1664
Query: 493 ALLYIVPFTLGKY 505
ALLY+VP TLG +
Sbjct: 1665ALLYLVPCTLGVF 1703
>TC77539 similar to GP|16118856|gb|AAL14629.1 growth-on protein GRO10
{Euphorbia esula}, partial (44%)
Length = 2020
Score = 351 bits (901), Expect = 4e-97
Identities = 170/220 (77%), Positives = 190/220 (86%)
Frame = +3
Query: 292 TCLTALLSCFRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQ 351
TCL ALLS RWF++ +++YVK+PF GA+ YLTLAVTPFCI FAV WAV R S++WIGQ
Sbjct: 3 TCLVALLS--RWFKHASESYVKLPFVGAISYLTLAVTPFCITFAVFWAVYRDKSFSWIGQ 176
Query: 352 DILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSG 411
DILGI LIITVLQIV + NLKVGTVLLSC+ LYD+ WVFVSK +F ESVMIVVARGD+SG
Sbjct: 177 DILGITLIITVLQIVHVSNLKVGTVLLSCSLLYDLFWVFVSKKFFKESVMIVVARGDRSG 356
Query: 412 EDGIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTA 471
EDGIPMLLK PR+ DPWGGYSIIGFGDI+LPG++VAFSLRYDWLAKK L +GYF+WAM A
Sbjct: 357 EDGIPMLLKFPRILDPWGGYSIIGFGDILLPGMLVAFSLRYDWLAKKTLVSGYFLWAMLA 536
Query: 472 YGLGLLITYVALNLMDGHGQPALLYIVPFTLGKYNHLTHK 511
YG GLLITYVALNLMDGHGQPALLYIVPFTLG L K
Sbjct: 537 YGFGLLITYVALNLMDGHGQPALLYIVPFTLGTIIALAQK 656
>TC84727 similar to GP|16118856|gb|AAL14629.1 growth-on protein GRO10
{Euphorbia esula}, partial (47%)
Length = 936
Score = 346 bits (888), Expect = 1e-95
Identities = 175/276 (63%), Positives = 213/276 (76%), Gaps = 1/276 (0%)
Frame = +2
Query: 15 VFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLE 74
+ L V VLAGDIVH DD P +PGCEN FVLVKV TW++GVE+AE+VGVGARFG T+
Sbjct: 110 LLLTKVPSVLAGDIVH-DDSTPKKPGCENQFVLVKVQTWVNGVEDAEFVGVGARFGRTIV 286
Query: 75 SKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIIN 134
SKEK+A TR+V++DP DCCS P NK+ ++I+V RG C+FT KAN A A ASAILIIN
Sbjct: 287 SKEKNARQTRLVLSDPRDCCSPPMNKIVGDVIMVDRGNCTFTKKANSAQNANASAILIIN 466
Query: 135 YRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAE 194
+ EL+KMVC+ +ETD+ I IPA MLP DAG LE +++ S VS+QLYSP RP VD+AE
Sbjct: 467 NQKELYKMVCDPDETDLSIHIPAGMLPLDAGTKLENMLKSTSSVSVQLYSPQRPTVDIAE 646
Query: 195 VFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYV-AESVGSRGYVEISTTAAI 253
VFLWLMAV TILCASYWSAW AREAA+E +KLL+D SDE + V G V ++ AA+
Sbjct: 647 VFLWLMAVLTILCASYWSAWRAREAAVEYDKLLRDVSDEISNTKDVAVSGVVNMNAKAAV 826
Query: 254 LFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEG 289
LFV++ASCFL MLYKLMS WF++VL LF IGGI+G
Sbjct: 827 LFVLVASCFLFMLYKLMSPWFIDVLXGLFXIGGIKG 934
>TC93164 similar to GP|16118854|gb|AAL14628.1 growth-on protein GRO11
{Euphorbia esula}, partial (28%)
Length = 774
Score = 264 bits (675), Expect = 6e-71
Identities = 126/132 (95%), Positives = 127/132 (95%)
Frame = +2
Query: 380 CAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDI 439
CAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDI
Sbjct: 2 CAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDI 181
Query: 440 ILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVP 499
ILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVP
Sbjct: 182 ILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVP 361
Query: 500 FTLGKYNHLTHK 511
FTLG + L K
Sbjct: 362 FTLGTFLSLGKK 397
>BF641501 similar to GP|16118854|gb| growth-on protein GRO11 {Euphorbia
esula}, partial (18%)
Length = 485
Score = 256 bits (653), Expect = 2e-68
Identities = 123/124 (99%), Positives = 123/124 (99%)
Frame = +2
Query: 1 MVSLGVIYSNIVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENA 60
MVSLGVIYSNIVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENA
Sbjct: 113 MVSLGVIYSNIVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENA 292
Query: 61 EYVGVGARFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKAN 120
EYVGVGARFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKC FTTKAN
Sbjct: 293 EYVGVGARFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCXFTTKAN 472
Query: 121 IADE 124
IADE
Sbjct: 473 IADE 484
>TC78773 similar to GP|17473842|gb|AAL38345.1 unknown protein {Arabidopsis
thaliana}, partial (90%)
Length = 1449
Score = 100 bits (249), Expect = 1e-21
Identities = 82/270 (30%), Positives = 128/270 (47%), Gaps = 13/270 (4%)
Frame = +1
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLE----VLVVLFCIGGIEGLQTCLTALLSCF- 301
+S A+ F + S L+ L+ L F + VL F + GI L L + F
Sbjct: 352 MSKEHAMRFPFVGSAMLLSLFLLFKFLSKDLVNTVLTGYFFVLGIVALSATLLPSIKRFL 531
Query: 302 --RW------FQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDI 353
W +++P V+I F + + T FC W R+ W+ +I
Sbjct: 532 PNHWNDDLIVWRFPYFRSVEIEFTRSQIVAAVPGTFFC-----AWYALRKH---WLANNI 687
Query: 354 LGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGED 413
LG+A I ++++ + + K G +LL+ F+YDI WVF + VM+ VA+ +
Sbjct: 688 LGLAFCIQGIEMLSLGSFKTGAILLAGLFVYDIFWVFFT------PVMVSVAKSFDA--- 840
Query: 414 GIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYG 473
P+ L P D +S++G GDI++PG+ VA +LR+D + + YF A Y
Sbjct: 841 --PIKLLFPTA-DSARPFSMLGLGDIVIPGIFVALALRFD--VSRGRKPQYFKSAFLGYT 1005
Query: 474 LGLLITYVALNLMDGHGQPALLYIVPFTLG 503
GL++T V +N QPALLYIVP +G
Sbjct: 1006FGLVLTIVVMNWFQA-AQPALLYIVPAVIG 1092
>TC77811 similar to GP|17473842|gb|AAL38345.1 unknown protein {Arabidopsis
thaliana}, partial (91%)
Length = 1284
Score = 85.5 bits (210), Expect = 5e-17
Identities = 78/279 (27%), Positives = 124/279 (43%), Gaps = 22/279 (7%)
Frame = +3
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLE----VLVVLFCIGGIEGLQTCLTALLSCFR 302
+S A+ F + S L+ L+ L F + VL + F + GI L L + F
Sbjct: 138 MSNEHAMRFPFVGSAMLLSLFLLFKFLSKDLVNTVLTLYFFVLGIVALSATLLPYIKRFL 317
Query: 303 ---W------FQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDI 353
W + +P ++I F + + T FC +A+ W+ +I
Sbjct: 318 PKPWNDDLIVWHFPYFRSLEIEFTKSQIIAAIPGTFFCGWYAL--------KKHWLANNI 473
Query: 354 LGIALIITVL---------QIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVV 404
LG+A I + +++ + + K G +LL F YDI WVF + VM+ V
Sbjct: 474 LGLAFCIQGICMGSPEEGIEMLSLGSFKTGAILLVGLFFYDIFWVFFTP------VMVSV 635
Query: 405 ARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGY 464
A+ + P+ L P D +S++G GDI++PG+ VA +LR+D + + Y
Sbjct: 636 AKSFDA-----PIKLLFPTA-DSKRPFSMLGLGDIVIPGIFVALALRFD--VSRGKQPQY 791
Query: 465 FVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTLG 503
F A Y G+ +T +N QPALLYIVP +G
Sbjct: 792 FKSAFLGYTFGIGLTIFVMNWFQA-AQPALLYIVPAVIG 905
>TC77812 similar to GP|17473842|gb|AAL38345.1 unknown protein {Arabidopsis
thaliana}, partial (74%)
Length = 809
Score = 71.6 bits (174), Expect = 7e-13
Identities = 61/220 (27%), Positives = 101/220 (45%), Gaps = 13/220 (5%)
Frame = +1
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLE----VLVVLFCIGGIEGLQTCLTALLSCFR 302
+S A+ F + S L+ L+ L F + VL + F + GI L L + F
Sbjct: 187 MSNEHAMRFPFVGSAMLLSLFLLFKFLSKDLVNTVLTLYFFVLGIVALSATLLPYIKRFL 366
Query: 303 ---W------FQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDI 353
W + +P ++I F + + T FC +A+ W+ +I
Sbjct: 367 PKPWNDDLIVWHFPYFRSLEIEFTKSQIIAAIPGTFFCGWYAL--------KKHWLANNI 522
Query: 354 LGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGED 413
LG+A I ++++ + + K G +LL F YDI WVF + VM+ VA+ +
Sbjct: 523 LGLAFCIQGIEMLSLGSFKTGAILLVGLFFYDIFWVFFT------PVMVSVAKSFDA--- 675
Query: 414 GIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYD 453
P+ L P D +S++G GDI++PG+ VA +LR+D
Sbjct: 676 --PIKLLFPTA-DSKRPFSMLGLGDIVIPGIFVALALRFD 786
>TC78359 similar to GP|16648809|gb|AAL25595.1 AT4g33410/F17M5_170
{Arabidopsis thaliana}, partial (95%)
Length = 1404
Score = 70.5 bits (171), Expect = 2e-12
Identities = 54/178 (30%), Positives = 85/178 (47%), Gaps = 13/178 (7%)
Frame = +1
Query: 241 SRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSC 300
S + + + A++ V++S LV+++ L S + T TA+ +
Sbjct: 319 SEASITLDRSQALMIPVMSSFSLVLMFYLFS--------------SVSQFLTAFTAIAAA 456
Query: 301 FRWFQY--PAQTYVKIPFFGAVPYLTL----------AVTPFCIVFAVV-WAVKRQASYA 347
F P Y+K F A PY++ A+ F F VV W V S
Sbjct: 457 SSLFFVLSPYAAYLKSQFGLADPYVSRCCSKSFTRIQAMLLFTCTFTVVAWLV----SGH 624
Query: 348 WIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVA 405
WI ++LGI++ I + VR+PN+K+ +LL C F+YDI WVF S+ +F +VM+ VA
Sbjct: 625 WILNNLLGISICIAFVSHVRLPNIKICAMLLLCLFVYDIFWVFYSERFFGANVMVSVA 798
Score = 48.9 bits (115), Expect = 5e-06
Identities = 26/83 (31%), Positives = 48/83 (57%), Gaps = 10/83 (12%)
Frame = +2
Query: 431 YSIIGFGDIILPGLVVAFSLRYDWLAKKNL---------RAGYFVW-AMTAYGLGLLITY 480
+ ++G GD+ +PG+++A L +D+ ++ + ++W A+ Y +GL +T
Sbjct: 953 FMMLGLGDMAIPGMLLALVLCFDYRKSRDTINLTDLHSSKGHKYIWYALPGYAIGL-VTA 1129
Query: 481 VALNLMDGHGQPALLYIVPFTLG 503
+A ++ QPALLY+VP TLG
Sbjct: 1130 LAAGVLSHSPQPALLYLVPSTLG 1198
>TC88456 similar to GP|15487292|dbj|BAB64531. vacuolar sorting receptor
{Vigna mungo}, partial (41%)
Length = 980
Score = 40.8 bits (94), Expect = 0.001
Identities = 26/84 (30%), Positives = 42/84 (49%), Gaps = 7/84 (8%)
Frame = +2
Query: 106 ILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDV-------DIGIPAV 158
+LV RG C FT KA A GA+AIL+ + R E + E +V I IP+
Sbjct: 443 VLVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNVVNDDYIEKINIPSA 622
Query: 159 MLPQDAGLNLERHIQNNSIVSIQL 182
++ + G +++ + + +V I L
Sbjct: 623 LISKSLGDRIKKALSDGEMVHINL 694
>TC87121 weakly similar to PIR|T06794|T06794 vacuolar sorting receptor
protein BP-80 - garden pea, partial (30%)
Length = 810
Score = 39.7 bits (91), Expect = 0.003
Identities = 27/84 (32%), Positives = 45/84 (53%), Gaps = 6/84 (7%)
Frame = +2
Query: 105 IILVHRGKCSFTTKANIADEAGASAILIINYRTE-LFKMVCEENETDVD-----IGIPAV 158
I+L+ RG+C F K A AGA+A+L+ + E L M E TD + I IP+V
Sbjct: 314 IVLLDRGECYFALKVWHAQLAGAAAVLVADSIDESLITMDSPEESTDAEGYIEKIVIPSV 493
Query: 159 MLPQDAGLNLERHIQNNSIVSIQL 182
++ + G +L+ + N V +++
Sbjct: 494 LVEKSFGDSLKEALNNKDEVLLRI 565
>TC77971 similar to PIR|T06794|T06794 vacuolar sorting receptor protein
BP-80 - garden pea, partial (97%)
Length = 2578
Score = 37.7 bits (86), Expect = 0.011
Identities = 24/84 (28%), Positives = 44/84 (51%), Gaps = 6/84 (7%)
Frame = +1
Query: 105 IILVHRGKCSFTTKANIADEAGASAILII-NYRTELFKMVCEENETD-----VDIGIPAV 158
I+L+ RG C F K A +AGAS++L+ + +L M E + +I IP+
Sbjct: 625 IVLLDRGSCFFALKVWNAQKAGASSVLVADDIEEKLITMDTPEEDGSSAKYIENITIPSA 804
Query: 159 MLPQDAGLNLERHIQNNSIVSIQL 182
++ ++ G L++ I +V++ L
Sbjct: 805 LIEKNFGEKLKKAISGGDMVNVNL 876
>TC92174 homologue to PIR|T06794|T06794 vacuolar sorting receptor protein
BP-80 - garden pea, partial (28%)
Length = 669
Score = 34.7 bits (78), Expect = 0.095
Identities = 38/149 (25%), Positives = 61/149 (40%), Gaps = 8/149 (5%)
Frame = +3
Query: 42 ENNFVLVKVPTWIDGVENAEYVGVGA-RFGPTLESKEKHANHTRVVMADPPDCCS-KPKN 99
E N + V P I G ++ G ++G ++ + D D S K K
Sbjct: 207 EKNSLSVTSPDKIKGKHDSAIGNFGIPQYGGSMAGNVVYPKDNNKGCKDFDDSSSFKSKP 386
Query: 100 KLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTE-LFKMVCEENETD-----VDI 153
I+L+ RG C F K A +AGASA+L+ + E L M E + +I
Sbjct: 387 GALPTILLLDRGSCFFALKVWNAXKAGASAVLVADDIEEPLITMDTPEEDVSSAKYIENI 566
Query: 154 GIPAVMLPQDAGLNLERHIQNNSIVSIQL 182
IP+ ++ + G L I + ++ L
Sbjct: 567 TIPSALIGKTFGXXLXDAIXGGDMXNVNL 653
>BQ147300 weakly similar to PIR|T01080|T01 hypothetical protein T10P11.3.1 -
Arabidopsis thaliana, partial (56%)
Length = 601
Score = 31.6 bits (70), Expect = 0.81
Identities = 25/116 (21%), Positives = 53/116 (45%)
Frame = +3
Query: 253 ILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYV 312
++ +++A F ++L +S ++ + V LF +G L T ++ F A +
Sbjct: 231 VIILIVAIIFTIILLFALSKYYKKHPVNLFLLG----LYTLCMSVAVGF------ACVFA 380
Query: 313 KIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRI 368
K P +LT V + F WAVKR ++++ + L++ + +++I
Sbjct: 381 KAPVVLEAAFLT-GVVVASLTFYTFWAVKRGKDFSFLAPFLFASLLVLMMFALIQI 545
>TC79025 similar to PIR|T49099|T49099 dihydrolipoamide S-acetyltransferase
precursor - Arabidopsis thaliana, partial (39%)
Length = 1257
Score = 31.2 bits (69), Expect = 1.1
Identities = 20/69 (28%), Positives = 33/69 (46%)
Frame = -3
Query: 229 DASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIE 288
DAS+ Y S + G + +L +L SC++V SFWF E+L++ F I
Sbjct: 1054 DASNPYSKISNFAAGLIRAVFVFFVLTSLLVSCWVV-----FSFWFTELLLIEFLTASIS 890
Query: 289 GLQTCLTAL 297
+ + A+
Sbjct: 889 LASSTVIAI 863
>TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycopersicon
esculentum}, partial (37%)
Length = 1230
Score = 30.8 bits (68), Expect = 1.4
Identities = 18/59 (30%), Positives = 32/59 (53%)
Frame = +3
Query: 118 KANIADEAGASAILIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNS 176
K +AG +A++++N E F + DV + +PAV + AGLN++ +I + S
Sbjct: 132 KGQAVKDAGGAAMILLNSEGEDFNPIA-----DVHV-LPAVHVSYSAGLNIQDYINSTS 290
>TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylase putative
{Arabidopsis thaliana}, partial (68%)
Length = 1310
Score = 30.8 bits (68), Expect = 1.4
Identities = 15/57 (26%), Positives = 26/57 (45%)
Frame = +2
Query: 294 LTALLSCFRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIG 350
L CF++F T F ++ L + FC ++ W +R+ ++AWIG
Sbjct: 116 LVCFFFCFKFFHQNVPTS-----FSSIVTLFFLLCFFCFFNSITWFGRRKQNHAWIG 271
>BF649032 weakly similar to PIR|T45951|T459 hypothetical protein F7J8.90 -
Arabidopsis thaliana, partial (11%)
Length = 387
Score = 30.0 bits (66), Expect = 2.3
Identities = 14/32 (43%), Positives = 17/32 (52%)
Frame = +3
Query: 273 WFLEVLVVLFCIGGIEGLQTCLTALLSCFRWF 304
WFL +L +LF GI G CL L RW+
Sbjct: 138 WFLILLFILFLSMGIVGTMMCLGRLKCGMRWW 233
>BF005832 similar to PIR|T05571|T055 hypothetical protein F22K18.170 -
Arabidopsis thaliana, partial (23%)
Length = 480
Score = 28.9 bits (63), Expect = 5.2
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 3/45 (6%)
Frame = +3
Query: 494 LLYIVP---FTLGKYNHLTHKFESVNTYGGILVFNVLFDCVLLVV 535
LL IVP F + HL H F S N ILV V+F+ +L++
Sbjct: 312 LLVIVPVIIFCVFVARHLRHHFSSYNAGYAILVVAVIFNIYVLIL 446
>BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (33%)
Length = 752
Score = 28.5 bits (62), Expect = 6.8
Identities = 13/33 (39%), Positives = 20/33 (60%)
Frame = +3
Query: 501 TLGKYNHLTHKFESVNTYGGILVFNVLFDCVLL 533
T Y+HLTH F+ +N+ I F ++ C+LL
Sbjct: 171 TFSFYHHLTHTFKEMNSISKI-AFPMILTCLLL 266
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,217,458
Number of Sequences: 36976
Number of extensions: 295576
Number of successful extensions: 2154
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 2119
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2143
length of query: 540
length of database: 9,014,727
effective HSP length: 101
effective length of query: 439
effective length of database: 5,280,151
effective search space: 2317986289
effective search space used: 2317986289
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146341.6


