

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146333.5 + phase: 0
(501 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
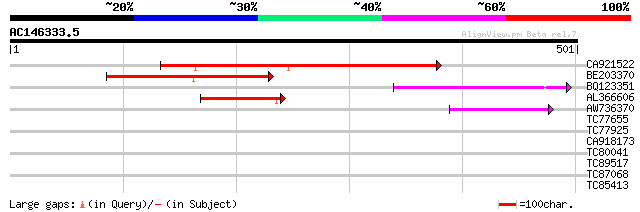
Score E
Sequences producing significant alignments: (bits) Value
CA921522 241 5e-64
BE203370 153 1e-37
BQ123351 101 6e-22
AL366606 100 2e-21
AW736370 44 2e-04
TC77655 similar to PIR|T09217|T09217 protein sam2B - spinach, pa... 31 1.3
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 30 1.7
CA918173 weakly similar to GP|11034537|dbj putative glucosyl tra... 30 2.8
TC80041 similar to GP|8778702|gb|AAF79710.1| T1N15.20 {Arabidops... 29 3.7
TC89517 weakly similar to PIR|A85475|A85475 hypothetical protein... 29 4.8
TC87068 similar to PIR|G96703|G96703 unknown protein 30164-3299... 28 6.3
TC85413 similar to PIR|S23737|S23737 proline-rich protein precur... 28 8.2
>CA921522
Length = 767
Score = 241 bits (615), Expect = 5e-64
Identities = 123/252 (48%), Positives = 162/252 (63%), Gaps = 4/252 (1%)
Frame = +3
Query: 134 KTSDVSKHLTTKSHIRGFTSKYLLEQANL--ETTCQDALEAILALLVYGLILFPNLDNFV 191
+T +V HL T+ GF++ +L E+ + A +ILALLVYGL+LFP+LDNFV
Sbjct: 3 ETDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFV 182
Query: 192 DMNAIEIFHSRNPVPTLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHLPSS--FHDN 249
D+ AI+IF SRNPVPTLL D Y +IH RT GR ILCC LLYRW SHLP + F N
Sbjct: 183 DIKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPRFTTN 362
Query: 250 SEDWSYSQRMMALSPNEVVWITPAAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQF 309
E+ +S+R+M+L+P EVVW II CG F NVPLLG GGI+YNP LA QF
Sbjct: 363 PENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARHQF 542
Query: 310 GFPMKTKPINLATSPEFFYYSNAPTGQREAFTRAWSKVRWKSVKHLGVRSGIAHEAYTQW 369
G+PM+ KP+++ ++ ++ TG RE RAW +R + LG ++G +YTQW
Sbjct: 543 GYPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSSYTQW 722
Query: 370 VINRAEEIGLPY 381
VI+RA +IG+PY
Sbjct: 723 VIDRAVQIGMPY 758
>BE203370
Length = 454
Score = 153 bits (387), Expect = 1e-37
Identities = 78/150 (52%), Positives = 105/150 (70%), Gaps = 2/150 (1%)
Frame = +1
Query: 86 CFTFPDFQLVPTLEAYSHLLDSPIAEKTPFTGPGASLTPLVIAKDLYLKTSDVSKHLTTK 145
CFTFPD+QL PTLE YS+L+ P+ +K PFTG IA L+L+TS + LT K
Sbjct: 4 CFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETSLIKAKLTIK 183
Query: 146 SHIRGFTSKYLLEQAN--LETTCQDALEAILALLVYGLILFPNLDNFVDMNAIEIFHSRN 203
++ +K+L +QA+ + DA +ILALL+YGL+LFPN+DN+VD++AI++F ++N
Sbjct: 184 GNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLTKN 363
Query: 204 PVPTLLADMYHAIHDRTLKGRVYILCCVPL 233
PVPTLLADMYH+IHDR GR IL C PL
Sbjct: 364 PVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>BQ123351
Length = 725
Score = 101 bits (252), Expect = 6e-22
Identities = 59/157 (37%), Positives = 93/157 (58%)
Frame = +2
Query: 340 FTRAWSKVRWKSVKHLGVRSGIAHEAYTQWVINRAEEIGLPYPAMRHVAASAPSIPLPLP 399
F +AW V + LG RS + +E+YT+WVI+RA + G+P+P R +++ S LP+
Sbjct: 8 FIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLPMT 187
Query: 400 PTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRREL 459
T+E Q LA +REK W+ RY EAE+ I TL G+ EQK HE L +++++++ L
Sbjct: 188 SKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDDLL 367
Query: 460 EEKDELLMQDSKRARGRRNFYARYCGSDSESESEDHP 496
+EKD+LL + + + R + + G DS+ E P
Sbjct: 368 QEKDKLLKKHITK-KQRMDSMDLFDGPDSDFED*SFP 475
>AL366606
Length = 393
Score = 100 bits (248), Expect = 2e-21
Identities = 55/81 (67%), Positives = 58/81 (70%), Gaps = 6/81 (7%)
Frame = -1
Query: 169 ALEAILALLVYGLILFPNLDNFVDMNAIEIFHSRNPVPTLLADMYHAIHDRTLKGRVYIL 228
A AI L GLILFPNLDNFVDMNAIE+FHS+NPVPTLLAD YHAIHDRTLKGR YIL
Sbjct: 390 APSAIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYIL 211
Query: 229 CCVPLL------YRWFISHLP 243
L +R F S LP
Sbjct: 210 WLHILFSNRVVYFRTFRSSLP 148
>AW736370
Length = 488
Score = 43.5 bits (101), Expect = 2e-04
Identities = 25/92 (27%), Positives = 44/92 (47%)
Frame = -2
Query: 389 ASAPSIPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLML 448
A P+ PLP+ T++ YQE L RE WK Y + + T+ G EQ+ +++
Sbjct: 487 AITPAPPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*K 308
Query: 449 KKELVKVRRELEEKDELLMQDSKRARGRRNFY 480
+ K+ ++E D L + R + R + +
Sbjct: 307 DVIIAKLNERIKENDAALDRIPGRKKRRMDLF 212
>TC77655 similar to PIR|T09217|T09217 protein sam2B - spinach, partial (64%)
Length = 1341
Score = 30.8 bits (68), Expect = 1.3
Identities = 16/48 (33%), Positives = 29/48 (60%), Gaps = 3/48 (6%)
Frame = +3
Query: 435 DGKDEQKTHENLMLKKELVKVRRELEEK---DELLMQDSKRARGRRNF 479
DG++E + + + +KKEL KV +E E+ +L+ K+A GR ++
Sbjct: 468 DGEEESEEEKRMRVKKELEKVAKEQAERRATAQLMYDLGKKAYGRGSY 611
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 30.4 bits (67), Expect = 1.7
Identities = 17/44 (38%), Positives = 24/44 (53%)
Frame = +3
Query: 450 KELVKVRRELEEKDELLMQDSKRARGRRNFYARYCGSDSESESE 493
KE+ + + E +E DS+ + RR Y + SDSESESE
Sbjct: 522 KEIKQKAKSESESEESKESDSELRKKRRKSYKKSRESDSESESE 653
>CA918173 weakly similar to GP|11034537|dbj putative glucosyl transferase
{Oryza sativa (japonica cultivar-group)}, partial (19%)
Length = 769
Score = 29.6 bits (65), Expect = 2.8
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = -3
Query: 72 LVHTLVQFYDPSFRCFTFPDFQ 93
L+HTLVQ + PS RCF FP+++
Sbjct: 113 LIHTLVQKW-PS*RCFPFPNYE 51
>TC80041 similar to GP|8778702|gb|AAF79710.1| T1N15.20 {Arabidopsis
thaliana}, partial (7%)
Length = 1621
Score = 29.3 bits (64), Expect = 3.7
Identities = 31/116 (26%), Positives = 46/116 (38%), Gaps = 11/116 (9%)
Frame = +3
Query: 392 PSIPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENLIMT-----LDGKDEQKT---H 443
PS L LP + +L +ESR +++ ENL+++ G QKT
Sbjct: 705 PSSVLSLPQERTQGILNNLRVESRFREVAGNGKQRDENLVLSGCHFDNGGDQNQKTVSDR 884
Query: 444 ENLMLKKELVKVRRELEEKDELLMQDSKRARGRRNFYARYCGSDSESESED---HP 496
ENL L++++V+ + CG D ESE ED HP
Sbjct: 885 ENLSLRQDIVRPVEAI------------------------CGGDLESEMEDQQNHP 980
>TC89517 weakly similar to PIR|A85475|A85475 hypothetical protein AT4g40080
[imported] - Arabidopsis thaliana, partial (46%)
Length = 1177
Score = 28.9 bits (63), Expect = 4.8
Identities = 23/87 (26%), Positives = 35/87 (39%), Gaps = 1/87 (1%)
Frame = -3
Query: 184 FPNLDNFVDMNAIEIFHSRNPVPTLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHLP 243
F L F++ N H NP+ ++H IH Y+ + L W I P
Sbjct: 764 FLPLPKFINSNQ-NFIHHHNPI------IFHQIH--------YLNNHLVLFTHWSIRFFP 630
Query: 244 SSFHDNSEDWSYS-QRMMALSPNEVVW 269
+SFH + S+S Q + N +W
Sbjct: 629 NSFHQGQQRISFS*QITIGKIRNSFLW 549
>TC87068 similar to PIR|G96703|G96703 unknown protein 30164-32998
[imported] - Arabidopsis thaliana, partial (24%)
Length = 711
Score = 28.5 bits (62), Expect = 6.3
Identities = 17/68 (25%), Positives = 30/68 (44%)
Frame = +2
Query: 399 PPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRE 458
PP++ + E WK ++ N + +D KD +L+KE + +R+
Sbjct: 185 PPSSSFNQSSERTKDKEENNNWKGSLDDKPNDLPLVDTKDNVPGVLYDLLQKEGLGLRKA 364
Query: 459 LEEKDELL 466
EKD+ L
Sbjct: 365 GHEKDQSL 388
>TC85413 similar to PIR|S23737|S23737 proline-rich protein precursor -
kidney bean, partial (56%)
Length = 1139
Score = 28.1 bits (61), Expect = 8.2
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +1
Query: 219 RTLKGRVYILCCVPLLYRWFI 239
R LK RVYI CC +L WF+
Sbjct: 883 RVLKFRVYIDCCFLVLRDWFL 945
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,037,359
Number of Sequences: 36976
Number of extensions: 228754
Number of successful extensions: 1296
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1279
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1293
length of query: 501
length of database: 9,014,727
effective HSP length: 100
effective length of query: 401
effective length of database: 5,317,127
effective search space: 2132167927
effective search space used: 2132167927
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146333.5


