

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.5 + phase: 0
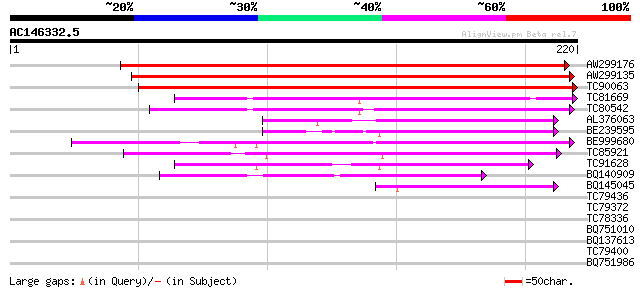
(220 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW299176 weakly similar to GP|12322903|gb unknown protein; 80797... 205 1e-53
AW299135 similar to PIR|T49964|T49 hypothetical protein F8M21.18... 176 6e-45
TC90063 weakly similar to GP|9759318|dbj|BAB09685.1 gb|AAF00668.... 151 2e-37
TC81669 weakly similar to GP|21739230|emb|CAD40985. OSJNBa0072F1... 77 4e-15
TC80542 similar to GP|18087595|gb|AAL58928.1 unknown protein {Ar... 69 1e-12
AL376063 weakly similar to PIR|H85044|H85 hypothetical protein A... 62 2e-10
BE239595 weakly similar to GP|21592507|gb| unknown {Arabidopsis ... 61 3e-10
BE999680 weakly similar to PIR|B71421|B714 hypothetical protein ... 60 7e-10
TC85921 weakly similar to GP|11908074|gb|AAG41466.1 unknown prot... 59 1e-09
TC91628 weakly similar to PIR|B71421|B71421 hypothetical protein... 53 8e-08
BQ140909 similar to GP|16973324|emb hypothetical protein {Gossyp... 51 3e-07
BQ145045 weakly similar to PIR|H85044|H850 hypothetical protein ... 44 4e-05
TC79436 similar to GP|18252183|gb|AAL61924.1 putative mitochondr... 32 0.25
TC79372 homologue to GP|2281647|gb|AAC49777.1| AP2 domain contai... 29 1.2
TC78336 similar to GP|21536509|gb|AAM60841.1 unknown {Arabidopsi... 29 1.6
BQ751010 similar to GP|9663769|emb| RNA Binding Protein 47 {Nico... 28 2.8
BQ137613 similar to PIR|S40770|S40 genome polyprotein - hepatiti... 28 3.6
TC79400 weakly similar to GP|20161447|dbj|BAB90371. hypothetical... 27 6.2
BQ751986 similar to PIR|T36527|T365 hypothetical protein SCH10.1... 27 6.2
>AW299176 weakly similar to GP|12322903|gb unknown protein; 80797-81587
{Arabidopsis thaliana}, partial (78%)
Length = 639
Score = 205 bits (522), Expect = 1e-53
Identities = 101/174 (58%), Positives = 129/174 (74%)
Frame = +1
Query: 44 TTKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDD 103
T P + GG KKG AI+DFILRLGAI AA+ AA MGT++Q LPFFTQF QF A +D
Sbjct: 49 TILVAPARPGGWKKGTAIMDFILRLGAIAAAIAAAASMGTSDQTLPFFTQFFQFEASYDS 228
Query: 104 FPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAA 163
F F+FFV++ AA +L+LSLPFSIV I+RP A GPR L+I+D V + L ++A++AA
Sbjct: 229 FTTFQFFVISMAIAASYLVLSLPFSIVAIIRPHAPGPRLFLIILDTVFLTLATSSAAAAA 408
Query: 164 AVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
++VYLAHNG+QD NW AIC QF DFC +S AVVASF+ V L LVV+S++A+
Sbjct: 409 SIVYLAHNGNQDTNWLAICNQFGDFCAQTSGAVVASFITVVVLILLVVMSALAI 570
>AW299135 similar to PIR|T49964|T49 hypothetical protein F8M21.180 -
Arabidopsis thaliana, partial (86%)
Length = 692
Score = 176 bits (446), Expect = 6e-45
Identities = 84/172 (48%), Positives = 123/172 (70%)
Frame = -3
Query: 48 TPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMF 107
T QKG MK+G++ILDFILRL AI A L +A+ MGT ++ LPFFTQF++F A +DD P
Sbjct: 654 TSTQKGNMKRGVSILDFILRLIAIVATLASAIAMGTTDESLPFFTQFVRFRANYDDLPTL 475
Query: 108 KFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVY 167
+FFVVA+ +G+LILSLP SI+ I+R A R + +I+D V++ L+ A +S+AA++VY
Sbjct: 474 RFFVVASAIVSGYLILSLPLSILHIIRSSAGMTRVIFIILDTVMLGLLTAGSSAAASIVY 295
Query: 168 LAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
LAH G++ ANW A CQQ+ FC+ S +++ SF+A L+++S++ L R
Sbjct: 294 LAHKGNRKANWFAFCQQYNSFCERISGSLIGSFIAIPLFIMLILLSALVLSR 139
>TC90063 weakly similar to GP|9759318|dbj|BAB09685.1
gb|AAF00668.1~gene_id:MBL20.8~similar to unknown protein
{Arabidopsis thaliana}, partial (56%)
Length = 765
Score = 151 bits (382), Expect = 2e-37
Identities = 74/170 (43%), Positives = 112/170 (65%)
Frame = +1
Query: 51 QKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFF 110
Q+ GMK+G++I+DFILR+ A + LG+A+ MGT +Q +PF T+F++F + D P F FF
Sbjct: 4 QEKGMKRGLSIMDFILRIFAAMSTLGSALSMGTAKQTMPFATRFVRFKVSFHDLPTFLFF 183
Query: 111 VVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAH 170
V AN G+L LSL S IVR ++ R LLV +D V+ L+ + AS+AAA+VY+AH
Sbjct: 184 VTANSIVCGYLALSLVLSFFHIVRTISVKSRILLVFLDTVMFGLLTSGASAAAAIVYVAH 363
Query: 171 NGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKRT 220
G+ ANW CQQ+ FC S ++V SF+A V L+++S +++ ++
Sbjct: 364 YGNPSANWFPFCQQYNSFCGRISGSLVGSFIAVVIFMILILMSGISISKS 513
>TC81669 weakly similar to GP|21739230|emb|CAD40985. OSJNBa0072F16.9 {Oryza
sativa}, partial (15%)
Length = 859
Score = 77.4 bits (189), Expect = 4e-15
Identities = 52/157 (33%), Positives = 83/157 (52%), Gaps = 1/157 (0%)
Frame = +3
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILS 124
ILR+ I + V+ TN Q + F+ ++F A + FKFFV ANG +L+
Sbjct: 174 ILRILVIVLTAVSIVVTVTNNQSVMLFS--IRFEAHFYYTSSFKFFVAANGVVCFMSVLT 347
Query: 125 LPFSIVCIVR-PLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQ 183
L F+++ + P FLL +VDLV+ L++A S+A AV Y+ G + W AIC
Sbjct: 348 LIFNLLMRQQTPQRKDYYFLLFLVDLVMTVLLIAGCSAATAVGYVGQYGEKHVGWTAICD 527
Query: 184 QFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKRT 220
FC+ + ++++ S++A F A L + +A K T
Sbjct: 528 HVQKFCKTNLISLLLSYLA--FFANLGLTILIAYKFT 632
>TC80542 similar to GP|18087595|gb|AAL58928.1 unknown protein {Arabidopsis
thaliana}, partial (66%)
Length = 1262
Score = 69.3 bits (168), Expect = 1e-12
Identities = 46/172 (26%), Positives = 84/172 (48%), Gaps = 7/172 (4%)
Frame = +3
Query: 55 MKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVAN 114
+ + + I + +LR +G + A V++ T+ Q+ FFT +Q A++ D F VVAN
Sbjct: 504 LDRRVRITELVLRFVNLGLGVLAVVLIVTDSQVREFFT--IQKKAKFTDMKALVFLVVAN 677
Query: 115 GAAAGFLILSLPFSIVCIVR-------PLAAGPRFLLVIVDLVLMALVVAAASSAAAVVY 167
AAG+ ++ +V +++ PLA + + D V+ + VAA S+AA
Sbjct: 678 AIAAGYSLIQGLRCVVSMIKGSVLFNKPLA----WAIFSCDQVMAYITVAAVSAAAQSAV 845
Query: 168 LAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
A G + W IC + FC + ++F+ S+ + + +S+ +L R
Sbjct: 846 FAKMGQEQLQWMKICNMYGKFCNQVGEGLASAFLVSLSMVVVSCISTFSLFR 1001
>AL376063 weakly similar to PIR|H85044|H85 hypothetical protein AT4g03540
[imported] - Arabidopsis thaliana, partial (73%)
Length = 504
Score = 62.0 bits (149), Expect = 2e-10
Identities = 38/117 (32%), Positives = 62/117 (52%), Gaps = 2/117 (1%)
Frame = +1
Query: 99 AQWDDFPMFKFFVVANGAAA--GFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVV 156
A++ + P FK+FV+AN GF +L LP + L+V +D+VL L++
Sbjct: 4 AKYTNSPAFKYFVIANSIVTVYGFFVLFLPAESLLWR---------LVVAMDMVLTMLLI 156
Query: 157 AAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
++ S+A A+ +A NG+ A W IC FC + A+VASF+ + L++ S
Sbjct: 157 SSISAALAIAQVAKNGNNYAAWLPICGSVPKFCNHMTGALVASFIGVIIYLILLLHS 327
>BE239595 weakly similar to GP|21592507|gb| unknown {Arabidopsis thaliana},
partial (44%)
Length = 651
Score = 61.2 bits (147), Expect = 3e-10
Identities = 38/123 (30%), Positives = 64/123 (51%), Gaps = 8/123 (6%)
Frame = +1
Query: 99 AQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRF--------LLVIVDLV 150
A++ P F +FVVANG I+SL ++ I + + GP+F L+ + D +
Sbjct: 184 AKFQHTPAFVYFVVANG------IVSL-HNLEMIAKHIL-GPKFHNKGLQLALIAVFDTM 339
Query: 151 LMALVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLV 210
+AL + +A A+ L NG+ A WN IC +F +C +++ASF+ + L +
Sbjct: 340 ALALASSGDGAATAMSELGRNGNSHAKWNKICDKFESYCNRGGGSLIASFIGLILLLIIT 519
Query: 211 VVS 213
V+S
Sbjct: 520 VMS 528
>BE999680 weakly similar to PIR|B71421|B714 hypothetical protein -
Arabidopsis thaliana, partial (32%)
Length = 616
Score = 60.1 bits (144), Expect = 7e-10
Identities = 46/203 (22%), Positives = 90/203 (43%), Gaps = 8/203 (3%)
Frame = +3
Query: 25 GKGKSIDGDHSPPHAATVVTTKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTN 84
G+GK+I + + K P G + + L F+L L AA+++GT+
Sbjct: 24 GRGKAIIDGVEGNYGRDELAMKKPPAGGGSCELVLXFLGFVL-------TLAAAIVVGTD 182
Query: 85 EQ--ILPFFTQF------LQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPL 136
+Q I+P + A+W F +F+VAN A + +S+ +++ +
Sbjct: 183 KQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSK 362
Query: 137 AAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAV 196
L+ I D +++AL+ + +A A+ L + G+ W +C F +C + ++
Sbjct: 363 VFWGT-LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASI 539
Query: 197 VASFVASVFLACLVVVSSVALKR 219
+ S + S+ LVV+ + R
Sbjct: 540 ILSQLGSLVFLLLVVLLPILRSR 608
>TC85921 weakly similar to GP|11908074|gb|AAG41466.1 unknown protein
{Arabidopsis thaliana}, partial (38%)
Length = 1032
Score = 58.9 bits (141), Expect = 1e-09
Identities = 43/173 (24%), Positives = 79/173 (44%), Gaps = 3/173 (1%)
Frame = +3
Query: 45 TKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFH--AQWD 102
+ +TP + + D ILR A++ + V+M T Q T +L A++
Sbjct: 228 SSSTPAPASSVGVDHSKFDLILRFLLFAASVASVVVMVTGNQ-----TVYLPVPRPAKFR 392
Query: 103 DFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFL-LVIVDLVLMALVVAAASS 161
P F +FV A A + I++ S+ I +P L L+ D V++ ++ +A +
Sbjct: 393 YSPAFVYFVAAFSVAGLYSIITTFISLSAIRKPNLKTKLLLHLIFWDAVMLGILASATGT 572
Query: 162 AAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSS 214
A +V YL G++ +W+ IC + FC+ +V S+ L+ +S+
Sbjct: 573 AGSVAYLGLKGNKHTDWHKICHIYDKFCRHIGASVGVGLFGSIVTLLLIWISA 731
>TC91628 weakly similar to PIR|B71421|B71421 hypothetical protein -
Arabidopsis thaliana, partial (30%)
Length = 731
Score = 53.1 bits (126), Expect = 8e-08
Identities = 41/152 (26%), Positives = 70/152 (45%), Gaps = 13/152 (8%)
Frame = +2
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQF--------LQFHAQWDDFPMFKFFVVANGA 116
+LRL A+ L AAV++ ++Q + + A+W + +FVVAN
Sbjct: 296 VLRLCALVLTLTAAVVLVADKQTTVVPVKISDSLPPLDVPVTAKWQYVSAYVYFVVANFI 475
Query: 117 AAGFLILSLPFSIVCIVRPLAAGPRF-----LLVIVDLVLMALVVAAASSAAAVVYLAHN 171
A + LS V LA G + L+ ++D +++AL+ + +A A+ LA
Sbjct: 476 AFAYATLSF-------VIALANGHKSKLLVTLVTLLDAIMVALLFSGNGAAWAIGVLAEK 634
Query: 172 GSQDANWNAICQQFTDFCQGSSLAVVASFVAS 203
G+ WN +C F FC ++ A + S + S
Sbjct: 635 GNSHVMWNKMCHVFDKFCNQAAAACLISLLGS 730
>BQ140909 similar to GP|16973324|emb hypothetical protein {Gossypium
hirsutum}, partial (47%)
Length = 534
Score = 51.2 bits (121), Expect = 3e-07
Identities = 35/127 (27%), Positives = 59/127 (45%)
Frame = +3
Query: 59 IAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAA 118
+ I++ LRL I + A +IM N + + + + D F++ V ANG A
Sbjct: 171 LRIVETFLRLFPICLCVTALIIMLXNSEENEYGS------VAYSDLGAFRYLVHANGICA 332
Query: 119 GFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANW 178
G+ + S IV + RP + ++D V +++AA + V+YLA G A W
Sbjct: 333 GYSLFSAV--IVAVPRPSTMPKAWAFFLLDQVQTYIILAAGAVLTEVLYLAEYGVPTAGW 506
Query: 179 NAICQQF 185
++ C F
Sbjct: 507 SSXCGSF 527
>BQ145045 weakly similar to PIR|H85044|H850 hypothetical protein AT4g03540
[imported] - Arabidopsis thaliana, partial (46%)
Length = 464
Score = 44.3 bits (103), Expect = 4e-05
Identities = 24/74 (32%), Positives = 41/74 (54%), Gaps = 3/74 (4%)
Frame = +1
Query: 143 LLVIVDL---VLMALVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVAS 199
L+ ++D VL L++++ S+A A+ +A NG+ A W IC FC + A+VAS
Sbjct: 109 LIALIDFSEQVLTMLLISSISAALAIAQVAKNGNNYAAWLPICGSVPKFCNHMTGALVAS 288
Query: 200 FVASVFLACLVVVS 213
F+ + L++ S
Sbjct: 289 FIGVIIYLILLLHS 330
>TC79436 similar to GP|18252183|gb|AAL61924.1 putative mitochondrial carrier
protein {Arabidopsis thaliana}, partial (77%)
Length = 1021
Score = 31.6 bits (70), Expect = 0.25
Identities = 17/51 (33%), Positives = 22/51 (42%)
Frame = +3
Query: 80 IMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIV 130
++ TN I P F Q WD KF+VV G G + P S+V
Sbjct: 132 VLITNCLINPMAQSFGQTEINWDKLDKTKFYVVGAGLFTGVTVALYPVSVV 284
>TC79372 homologue to GP|2281647|gb|AAC49777.1| AP2 domain containing
protein RAP2.11 {Arabidopsis thaliana}, partial (27%)
Length = 1086
Score = 29.3 bits (64), Expect = 1.2
Identities = 19/48 (39%), Positives = 29/48 (59%)
Frame = -3
Query: 121 LILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYL 168
L+L P ++V ++ PLA + ++V LVL +LVV+ SA VV L
Sbjct: 421 LVLVTPLALVLLIAPLAVP---IQLVVLLVLPSLVVSLVVSAWLVVLL 287
>TC78336 similar to GP|21536509|gb|AAM60841.1 unknown {Arabidopsis
thaliana}, partial (83%)
Length = 1728
Score = 28.9 bits (63), Expect = 1.6
Identities = 11/41 (26%), Positives = 28/41 (67%)
Frame = +3
Query: 122 ILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSA 162
+L L + +C++R L A RF+++++ L+ + L++ + ++A
Sbjct: 282 MLELFLNSLCLIRVLIANIRFIVILIALLRVVLILLSIAAA 404
>BQ751010 similar to GP|9663769|emb| RNA Binding Protein 47 {Nicotiana
plumbaginifolia}, partial (3%)
Length = 399
Score = 28.1 bits (61), Expect = 2.8
Identities = 15/29 (51%), Positives = 22/29 (75%)
Frame = +2
Query: 134 RPLAAGPRFLLVIVDLVLMALVVAAASSA 162
RP G R+LLV++ L+L+ LV AAA++A
Sbjct: 224 RPKHVGRRWLLVMLLLLLLLLVHAAAAAA 310
>BQ137613 similar to PIR|S40770|S40 genome polyprotein - hepatitis C virus,
partial (0%)
Length = 964
Score = 27.7 bits (60), Expect = 3.6
Identities = 27/93 (29%), Positives = 36/93 (38%)
Frame = -3
Query: 124 SLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQ 183
+LP I+CI+RPL PR L V L VV+ +A G W
Sbjct: 410 ALPVLILCIIRPLF--PRCCLFDVYLCFTFFVVSGLFRSA-----DRGGRASVGWGCFV- 255
Query: 184 QFTDFCQGSSLAVVASFVASVFLACLVVVSSVA 216
F G + + SF+ S F V SV+
Sbjct: 254 ----FLPGGTSFLCLSFLLSAFFFRCVFRGSVS 168
>TC79400 weakly similar to GP|20161447|dbj|BAB90371. hypothetical
protein~similar to Arabidopsis thaliana chromosome 1
F8K7.2, partial (12%)
Length = 1251
Score = 26.9 bits (58), Expect = 6.2
Identities = 17/54 (31%), Positives = 25/54 (45%), Gaps = 2/54 (3%)
Frame = -3
Query: 106 MFKFFVVANGAA--AGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVA 157
+F FFV A + +L LPF+ C + G R +++VD L V A
Sbjct: 736 LFAFFVKTPSATRTSSMELLELPFAFSCFTPFVGLGTRTAVLVVDTDLFVEVEA 575
>BQ751986 similar to PIR|T36527|T365 hypothetical protein SCH10.15 -
Streptomyces coelicolor, partial (10%)
Length = 603
Score = 26.9 bits (58), Expect = 6.2
Identities = 12/29 (41%), Positives = 19/29 (65%)
Frame = +1
Query: 189 CQGSSLAVVASFVASVFLACLVVVSSVAL 217
CQ L + +SFV S+ + C + +SSVA+
Sbjct: 481 CQLPGLFLGSSFVTSLVIVCAIGISSVAV 567
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,185,885
Number of Sequences: 36976
Number of extensions: 80642
Number of successful extensions: 598
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 577
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 587
length of query: 220
length of database: 9,014,727
effective HSP length: 92
effective length of query: 128
effective length of database: 5,612,935
effective search space: 718455680
effective search space used: 718455680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146332.5


