

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.3 - phase: 0
(281 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
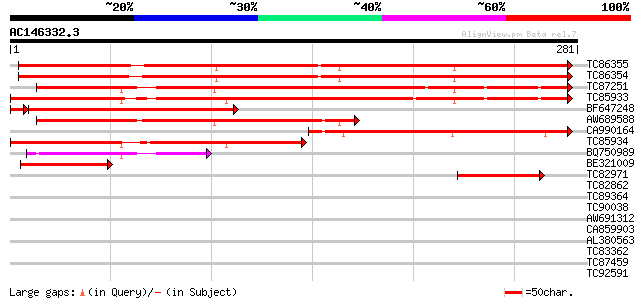
Score E
Sequences producing significant alignments: (bits) Value
TC86355 weakly similar to GP|9757805|dbj|BAB08323.1 CCR4-associa... 345 1e-95
TC86354 weakly similar to GP|9757805|dbj|BAB08323.1 CCR4-associa... 334 2e-92
TC87251 similar to GP|20197620|gb|AAD15397.2 putative CCR4-assoc... 239 9e-64
TC85933 similar to GP|20197620|gb|AAD15397.2 putative CCR4-assoc... 228 3e-60
BF647248 similar to PIR|F86293|F862 hypothetical protein AAF1848... 220 7e-58
AW689588 similar to GP|9757805|dbj CCR4-associated factor-like p... 174 4e-44
CA990164 similar to GP|9757805|dbj CCR4-associated factor-like p... 145 2e-35
TC85934 similar to GP|15292843|gb|AAK92792.1 putative CCR4-assoc... 121 3e-28
BQ750989 weakly similar to PIR|T41149|T411 probable trascription... 75 2e-14
BE321009 weakly similar to GP|19171001|emb SIMILAR TO CCR4-ASSOC... 55 2e-08
TC82971 similar to PIR|T49142|T49142 CCR4-associated factor 1-li... 52 2e-07
TC82862 similar to PIR|T02323|T02323 nodulin-like protein [impor... 30 1.4
TC89364 weakly similar to PIR|C96745|C96745 hypothetical protein... 30 1.4
TC90038 similar to PIR|A84451|A84451 probable AAA-type ATPase [i... 29 1.8
AW691312 similar to GP|14586363|emb lateral root primordia (LRP1... 28 5.1
CA859903 27 6.7
AL380563 similar to GP|15450498|gb| AT5g02940/F9G14_250 {Arabido... 27 6.7
TC83362 27 6.7
TC87459 similar to GP|14596225|gb|AAK68840.1 Unknown protein {Ar... 27 8.8
TC92591 weakly similar to GP|21593452|gb|AAM65419.1 unknown {Ara... 27 8.8
>TC86355 weakly similar to GP|9757805|dbj|BAB08323.1 CCR4-associated
factor-like protein {Arabidopsis thaliana}, partial
(84%)
Length = 1041
Score = 345 bits (885), Expect = 1e-95
Identities = 178/278 (64%), Positives = 209/278 (75%), Gaps = 3/278 (1%)
Frame = +3
Query: 5 KEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHL 64
K KPI IRQVWA NLE EF LIR ++ +P++SMDTEFPGV+ +PN D +
Sbjct: 156 KMNQLKEKPIIIRQVWASNLEVEFALIRQVINQYPFISMDTEFPGVIYSPNVDRRL---- 323
Query: 65 RHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPG-DVAYSYIWEFNFKDFDVDRDLQ 123
+ PS+ Y +LK NVD L LIQ+G+TL+D NGNLP SYIWEFNF DFD +RDL
Sbjct: 324 --LKPSDHYRYLKVNVDALKLIQVGITLSDGNGNLPHFGTNNSYIWEFNFCDFDFERDLY 497
Query: 124 NPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDFGYLVK 182
N DSI++L RQGIDFKRNL +GVDS FA+ F L SGLV N V WVTFHS+YDFGYLVK
Sbjct: 498 NQDSIDMLCRQGIDFKRNLCHGVDSSRFAE-FMLTSGLVFNKSVVWVTFHSAYDFGYLVK 674
Query: 183 ILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGNSH 241
ILT+ LP+RLE+FL+ILT +FG+NVYDMK+M++FCN LYGGLERVAT L V R G SH
Sbjct: 675 ILTRRNLPNRLEDFLNILTILFGKNVYDMKHMMRFCNALYGGLERVATTLNVGRVAGKSH 854
Query: 242 QAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
QA SDSLLTW AFKKM D YF+NN HAGVLFGLE+
Sbjct: 855 QAGSDSLLTWHAFKKMMDTYFMNNEAQKHAGVLFGLEI 968
>TC86354 weakly similar to GP|9757805|dbj|BAB08323.1 CCR4-associated
factor-like protein {Arabidopsis thaliana}, partial
(76%)
Length = 1029
Score = 334 bits (857), Expect = 2e-92
Identities = 174/278 (62%), Positives = 208/278 (74%), Gaps = 3/278 (1%)
Frame = +1
Query: 5 KEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHL 64
K KPI IRQVWA NLE EFDLIR V+ +P++SMDTEFPGV+ +P D
Sbjct: 76 KMNQLKEKPIIIRQVWASNLEVEFDLIRQVVNQYPFISMDTEFPGVIYSPKVDRC----- 240
Query: 65 RHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPG-DVAYSYIWEFNFKDFDVDRDLQ 123
++ PS+ Y +LK NVD L LIQ+G+TL++ NGNLP YIWEFNF DFD + DL
Sbjct: 241 -NLKPSDHYRYLKVNVDALKLIQVGITLSNGNGNLPHFGTNNRYIWEFNFCDFDFEHDLY 417
Query: 124 NPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDFGYLVK 182
N DSI++L RQGIDFKRN +GV+S FA+ F L SGLV N V WVTFHS+YDFGYLVK
Sbjct: 418 NQDSIDMLCRQGIDFKRNFSHGVNSSRFAE-FMLTSGLVFNKSVVWVTFHSAYDFGYLVK 594
Query: 183 ILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGNSH 241
ILT+ LP+RLE+FL+ILT +FG+NVYDMK+M++FCN LYGGLERVA+ L V RAVG SH
Sbjct: 595 ILTRRNLPNRLEDFLNILTILFGKNVYDMKHMMRFCNALYGGLERVASTLNVCRAVGKSH 774
Query: 242 QAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
QA SDSLLTW AFKKM D +F+NN HAGVLFGLE+
Sbjct: 775 QAGSDSLLTWHAFKKMMDTHFLNNEAQKHAGVLFGLEI 888
>TC87251 similar to GP|20197620|gb|AAD15397.2 putative CCR4-associated
factor {Arabidopsis thaliana}, partial (96%)
Length = 1089
Score = 239 bits (610), Expect = 9e-64
Identities = 130/271 (47%), Positives = 182/271 (66%), Gaps = 5/271 (1%)
Frame = +2
Query: 14 IKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRHMDPSE 71
I+IR+VW++NLE EF +IR+ V +P+++MDTEFPG+V+ P NF N YH
Sbjct: 248 IQIREVWSDNLEEEFAVIREIVDDYPFIAMDTEFPGIVLRPVGNFKSNYDYH-------- 403
Query: 72 QYSFLKANVDNLNLIQLGLTLTDANGNLP--GDVAYSYIWEFNFKDFDVDRDLQNPDSIE 129
Y LK NVD L LIQLGLT +D +GNLP G+ IW+FNF++F+V+ D+ DSIE
Sbjct: 404 -YQTLKDNVDMLKLIQLGLTFSDEHGNLPTCGEDDRFCIWQFNFREFNVNEDVFANDSIE 580
Query: 130 LLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNYL 189
LLR+ GIDFK+N G+D+ F +L ++N V W+TFHS YDFGYL+K++T L
Sbjct: 581 LLRQSGIDFKKNNEDGIDARRFGELLMSSGIVLNDNVHWITFHSGYDFGYLLKLVTCQNL 760
Query: 190 PSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGNSHQAASDSL 248
P F +++ F +YD+K+++KFCN L+GGL ++A L+V R VG HQA SDSL
Sbjct: 761 PDTQVGFFNLINMYFPM-LYDIKHLMKFCNSLHGGLNKLAELLEVKR-VGICHQAGSDSL 934
Query: 249 LTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
LT AF+K+K+ +F + + +AGVL+GL V
Sbjct: 935 LTSSAFRKLKENFF-SGSLEKYAGVLYGLGV 1024
>TC85933 similar to GP|20197620|gb|AAD15397.2 putative CCR4-associated
factor {Arabidopsis thaliana}, partial (94%)
Length = 1362
Score = 228 bits (580), Expect = 3e-60
Identities = 135/283 (47%), Positives = 179/283 (62%), Gaps = 4/283 (1%)
Frame = +3
Query: 1 MTKVKEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDP 58
MT + ++ I+IR+VW +NLE EF LIR+ V + YV+MDTEFPGVV+ P NF
Sbjct: 291 MTDLPKDLQKGDSIQIREVWNDNLEEEFVLIREIVDKYNYVAMDTEFPGVVLRPVGNFK- 467
Query: 59 NIPYHLRHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYS-YIWEFNFKDFD 117
H+ + Y LK NVD L LIQLGLT +D NGNLP S IW+FNF++F+
Sbjct: 468 ----HINDFN----YQTLKDNVDMLKLIQLGLTFSDENGNLPTCGTDSPCIWQFNFREFN 623
Query: 118 VDRDLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDF 177
V D+ DSIELLR+ GIDFK+N G+D F +L ++N V WVTFHS YDF
Sbjct: 624 VSEDIFAADSIELLRQCGIDFKKNSEQGIDVNRFGELLMSSGIVLNDNVHWVTFHSGYDF 803
Query: 178 GYLVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRA 236
GYL+K+LT LP F ++ I+ VYD+K+++KFCN L+GGL ++A L V R
Sbjct: 804 GYLLKLLTCRALPDTQAGFFDLI-GIYFPIVYDIKHLMKFCNSLHGGLNKLAELLDVER- 977
Query: 237 VGNSHQAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
VG HQA SDSLLT F+K+++ +F N ++GVL+GL V
Sbjct: 978 VGVCHQAGSDSLLTACTFRKLRETFF-NGETEKYSGVLYGLGV 1103
>BF647248 similar to PIR|F86293|F862 hypothetical protein AAF18489.1
[imported] - Arabidopsis thaliana, partial (13%)
Length = 436
Score = 220 bits (560), Expect(2) = 7e-58
Identities = 103/104 (99%), Positives = 104/104 (99%)
Frame = +3
Query: 10 SNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDP 69
SNKPIKIRQVWAENL+REFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDP
Sbjct: 123 SNKPIKIRQVWAENLQREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDP 302
Query: 70 SEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNF 113
SEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNF
Sbjct: 303 SEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNF 434
Score = 21.6 bits (44), Expect(2) = 7e-58
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +2
Query: 1 MTKVKEEDA 9
MTKVKEEDA
Sbjct: 95 MTKVKEEDA 121
>AW689588 similar to GP|9757805|dbj CCR4-associated factor-like protein
{Arabidopsis thaliana}, partial (46%)
Length = 527
Score = 174 bits (440), Expect = 4e-44
Identities = 96/165 (58%), Positives = 117/165 (70%), Gaps = 5/165 (3%)
Frame = +2
Query: 14 IKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPSEQY 73
I IR VW+ NLE EF LIR FV P +SMDTEFPGVVV P+ ++ + H P+ Y
Sbjct: 29 ISIRPVWSFNLESEFKLIRSFVDSHPIISMDTEFPGVVVRPDGITDLTSY--HRTPATHY 202
Query: 74 SFLKANVDNLNLIQLGLTLTDANGNLP----GDVAYSYIWEFNFKDFDVDRDLQNPDSIE 129
S LKANVD LNLIQ+GLTL+DA GNLP G+ IWEFNF DFDV RD+ +SIE
Sbjct: 203 SVLKANVDGLNLIQVGLTLSDAKGNLPKLENGNSEEFLIWEFNFSDFDVVRDIHAHESIE 382
Query: 130 LLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHS 173
LL+ QGIDF++N +GV+S++FA+L + SGLV N VSWVTFHS
Sbjct: 383 LLKSQGIDFEKNKEFGVESMKFAELM-MSSGLVCNEEVSWVTFHS 514
>CA990164 similar to GP|9757805|dbj CCR4-associated factor-like protein
{Arabidopsis thaliana}, partial (45%)
Length = 731
Score = 145 bits (365), Expect = 2e-35
Identities = 75/134 (55%), Positives = 101/134 (74%), Gaps = 3/134 (2%)
Frame = +2
Query: 149 LEFAKLFRLKSGLVNS-GVSWVTFHSSYDFGYLVKILTQNYLPSRLEEFLSILTQIFGQN 207
++FA+L + SGLV + VSWVTFHS+YDFGYLVK+LTQ LP LEEFL ++ FG
Sbjct: 2 VKFAELM-MSSGLVCADSVSWVTFHSAYDFGYLVKLLTQRLLPDDLEEFLRLVKVFFGDK 178
Query: 208 VYDMKYMIKFC-NLYGGLERVATKLKVSRAVGNSHQAASDSLLTWQAFKKMKDIYFVN-N 265
V+D+K++++FC NL+GGL+RV LKV R +G SHQA SDSLLT AF+ ++++YF +
Sbjct: 179 VFDVKHLMRFCTNLHGGLDRVCRSLKVERLIGKSHQAGSDSLLTLHAFQNIRELYFGKAD 358
Query: 266 GITMHAGVLFGLEV 279
G +AGVL+GLEV
Sbjct: 359 GFVKYAGVLYGLEV 400
>TC85934 similar to GP|15292843|gb|AAK92792.1 putative CCR4-associated
factor {Arabidopsis thaliana}, partial (50%)
Length = 721
Score = 121 bits (304), Expect = 3e-28
Identities = 73/150 (48%), Positives = 95/150 (62%), Gaps = 3/150 (2%)
Frame = +3
Query: 1 MTKVKEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDP 58
MT + ++ I+IR+VW +NLE EF LIR+ V + YV+MDTEFPGVV+ P NF
Sbjct: 270 MTDLPKDLQKGDSIQIREVWNDNLEEEFVLIREIVDKYNYVAMDTEFPGVVLRPVGNF-- 443
Query: 59 NIPYHLRHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYS-YIWEFNFKDFD 117
+H++ Y LK NVD L LIQLGLT +D NGNLP S IW+FNF++F+
Sbjct: 444 ------KHIN-DFNYQTLKDNVDMLKLIQLGLTFSDENGNLPTCGTDSPCIWQFNFREFN 602
Query: 118 VDRDLQNPDSIELLRRQGIDFKRNLIYGVD 147
V D+ DSIELLR+ GI F +N G+D
Sbjct: 603 VSEDIFAADSIELLRQCGI*F*KNSEQGID 692
>BQ750989 weakly similar to PIR|T41149|T411 probable trascription factor
ccr4-associated factor homolog - fission yeast, partial
(22%)
Length = 668
Score = 75.5 bits (184), Expect = 2e-14
Identities = 42/94 (44%), Positives = 54/94 (56%), Gaps = 2/94 (2%)
Frame = +1
Query: 9 ASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRH 66
A NK KIR+VW NL E ++RD + +PY++MDTEFPGVV P +F YH
Sbjct: 274 AQNKG-KIREVWKHNLHEEMAVLRDLIEKYPYIAMDTEFPGVVSRPMGSFRGKSDYH--- 441
Query: 67 MDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLP 100
Y L+ NVD L +IQ+GLTL +G P
Sbjct: 442 ------YQCLRTNVDMLKVIQIGLTLFHEDGKTP 525
>BE321009 weakly similar to GP|19171001|emb SIMILAR TO CCR4-ASSOCIATED FACTOR
1 {Encephalitozoon cuniculi}, partial (12%)
Length = 302
Score = 55.5 bits (132), Expect = 2e-08
Identities = 24/46 (52%), Positives = 32/46 (69%)
Frame = +2
Query: 6 EEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVV 51
+ED +KPI IR+VW NL EF LI + + ++SMDTEFPG+V
Sbjct: 158 KEDPGSKPIMIRKVWGYNLSCEFKLISQLIGKYNFISMDTEFPGIV 295
>TC82971 similar to PIR|T49142|T49142 CCR4-associated factor 1-like protein
- Arabidopsis thaliana, partial (11%)
Length = 296
Score = 52.4 bits (124), Expect = 2e-07
Identities = 24/43 (55%), Positives = 31/43 (71%)
Frame = +2
Query: 223 GLERVATKLKVSRAVGNSHQAASDSLLTWQAFKKMKDIYFVNN 265
GL+RV L V R G SHQA SDSLLT AF+K+K+++F N+
Sbjct: 2 GLDRVGKTLNVDRVAGKSHQAGSDSLLTLHAFRKIKEVHFGND 130
>TC82862 similar to PIR|T02323|T02323 nodulin-like protein [imported] -
Arabidopsis thaliana, partial (5%)
Length = 792
Score = 29.6 bits (65), Expect = 1.4
Identities = 23/90 (25%), Positives = 46/90 (50%), Gaps = 1/90 (1%)
Frame = +3
Query: 115 DFDVDRDLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFR-LKSGLVNSGVSWVTFHS 173
D + D + S +LR + +DF +L+ G+D+ + LF+ LK L + GV
Sbjct: 90 DSSMQIDASSDVSQSVLRARLLDFAVSLLPGLDTKDIDLLFQVLKPALQDVGVMQ---KK 260
Query: 174 SYDFGYLVKILTQNYLPSRLEEFLSILTQI 203
+Y ++ + +++ S+LE L ++ +I
Sbjct: 261 AYKVLSIILRSSDSFVSSKLEVLLGLMVEI 350
>TC89364 weakly similar to PIR|C96745|C96745 hypothetical protein T9N14.3
[imported] - Arabidopsis thaliana, partial (45%)
Length = 1903
Score = 29.6 bits (65), Expect = 1.4
Identities = 17/62 (27%), Positives = 31/62 (49%), Gaps = 1/62 (1%)
Frame = +1
Query: 85 LIQLGLTLTDANGNLPGDVAY-SYIWEFNFKDFDVDRDLQNPDSIELLRRQGIDFKRNLI 143
L+ L L L +GN+P V+ S + N + + PD++E ++ +DF +N +
Sbjct: 358 LVDLNLALNSLSGNIPNSVSLMSSLNSLNLSRNKLTGTI--PDNLEKMKLSSVDFSQNSL 531
Query: 144 YG 145
G
Sbjct: 532 SG 537
>TC90038 similar to PIR|A84451|A84451 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, partial (43%)
Length = 1000
Score = 29.3 bits (64), Expect = 1.8
Identities = 23/97 (23%), Positives = 47/97 (47%), Gaps = 7/97 (7%)
Frame = +2
Query: 180 LVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCNLYGGLE-----RVATKLKVS 234
++K+ Q+ + +F S + ++ D+KY+ CN Y G + A K +
Sbjct: 674 ILKLKLQSLMKRNAFKF*SCIQGKIPRDSQDLKYVAASCNGYVGADLRDLCLEAVKSAIE 853
Query: 235 RAVGNSHQAASDSLLTWQAFKKMKDIY--FVNNGITM 269
R+ N+++ +DS LT + +K + + + GIT+
Sbjct: 854 RS-DNANKDVNDSSLTMEDWKNARSLVQPSITRGITV 961
>AW691312 similar to GP|14586363|emb lateral root primordia (LRP1)
{Arabidopsis thaliana}, partial (14%)
Length = 652
Score = 27.7 bits (60), Expect = 5.1
Identities = 12/28 (42%), Positives = 17/28 (59%)
Frame = -1
Query: 153 KLFRLKSGLVNSGVSWVTFHSSYDFGYL 180
KL ++KSGL V WV F +DF ++
Sbjct: 178 KLMKVKSGLNCDHVKWVGFGFGFDFDFV 95
>CA859903
Length = 569
Score = 27.3 bits (59), Expect = 6.7
Identities = 19/72 (26%), Positives = 33/72 (45%), Gaps = 6/72 (8%)
Frame = +3
Query: 31 IRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPS------EQYSFLKANVDNLN 84
+++ + FPY + P VV P+F P P H R+ PS ++ ++A V +
Sbjct: 78 LKNLIEHFPY*KIHYLTPPYVVVPDFLP--PLHPRNFRPSVDICETDKSYMVEAEVPGMK 251
Query: 85 LIQLGLTLTDAN 96
L + T+ N
Sbjct: 252 KDDLSVEFTNDN 287
>AL380563 similar to GP|15450498|gb| AT5g02940/F9G14_250 {Arabidopsis
thaliana}, partial (10%)
Length = 501
Score = 27.3 bits (59), Expect = 6.7
Identities = 13/44 (29%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Frame = +1
Query: 177 FGYLVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYM-IKFCN 219
FGY + ++ Y S L F+SIL+ +F + + + + + +K+ N
Sbjct: 307 FGYCIGVIDC*YCCSYLSSFMSILSHLFSE*IGEKRCLNVKYVN 438
>TC83362
Length = 252
Score = 27.3 bits (59), Expect = 6.7
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = -3
Query: 112 NFKDFDVDRDLQNPDSIELLR 132
N+ F D+D +NPD IELLR
Sbjct: 79 NYTMF*FDQDNENPDEIELLR 17
>TC87459 similar to GP|14596225|gb|AAK68840.1 Unknown protein {Arabidopsis
thaliana}, partial (9%)
Length = 2880
Score = 26.9 bits (58), Expect = 8.8
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Frame = -2
Query: 154 LFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNYLPSRLEE--FLSILT 201
L L VN ++W SS G ++ L +N+LPSR++ F+SI T
Sbjct: 2669 LSNLTINSVNICLAWQYDSSSTIQGSMIPSLFRNFLPSRIKTIYFISIYT 2520
>TC92591 weakly similar to GP|21593452|gb|AAM65419.1 unknown {Arabidopsis
thaliana}, partial (40%)
Length = 654
Score = 26.9 bits (58), Expect = 8.8
Identities = 16/48 (33%), Positives = 25/48 (51%)
Frame = +3
Query: 186 QNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCNLYGGLERVATKLKV 233
+N P L EF L N +M+ + +FCN+Y L+ + T+ KV
Sbjct: 78 KNAQPDPLGEFAENLVAEI--NTINMEDITRFCNVYVDLDFLVTEAKV 215
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,423,263
Number of Sequences: 36976
Number of extensions: 111592
Number of successful extensions: 552
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 530
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 532
length of query: 281
length of database: 9,014,727
effective HSP length: 95
effective length of query: 186
effective length of database: 5,502,007
effective search space: 1023373302
effective search space used: 1023373302
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146332.3


