

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.11 - phase: 0
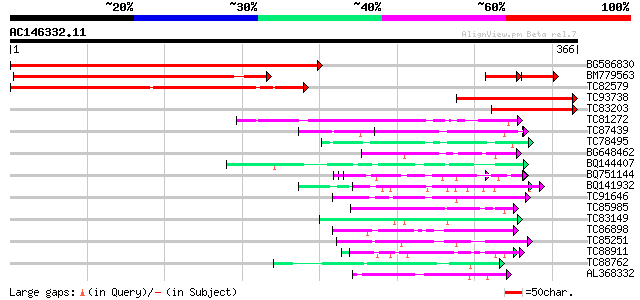
(366 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG586830 similar to PIR|T52141|T52 hypothetical protein [importe... 402 e-112
BM779563 similar to PIR|T52141|T52 hypothetical protein [importe... 286 7e-78
TC82579 similar to PIR|T52141|T52141 hypothetical protein [impor... 262 1e-70
TC93738 similar to GP|23617133|dbj|BAC20813. transcription facto... 164 5e-41
TC83203 78 6e-15
TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 ... 50 1e-06
TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in... 50 1e-06
TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 ... 49 3e-06
BG648462 weakly similar to GP|14253163|em VMP4 protein {Volvox c... 47 9e-06
BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 47 9e-06
BQ751144 weakly similar to PIR|PQ0479|PQ04 pistil extensin-like ... 47 9e-06
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 47 2e-05
TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 47 2e-05
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 46 2e-05
TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk p... 46 2e-05
TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrat... 46 3e-05
TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean... 45 4e-05
TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2... 45 4e-05
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 45 6e-05
AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein... 44 8e-05
>BG586830 similar to PIR|T52141|T52 hypothetical protein [imported] -
Arabidopsis thaliana, partial (46%)
Length = 699
Score = 402 bits (1033), Expect = e-112
Identities = 202/202 (100%), Positives = 202/202 (100%)
Frame = +1
Query: 1 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 60
MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG
Sbjct: 55 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 234
Query: 61 SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNA 120
SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNA
Sbjct: 235 SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNA 414
Query: 121 RECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVA 180
RECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVA
Sbjct: 415 RECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVA 594
Query: 181 DVPDDPSFINFFSAAMGIGNTS 202
DVPDDPSFINFFSAAMGIGNTS
Sbjct: 595 DVPDDPSFINFFSAAMGIGNTS 660
Score = 32.3 bits (72), Expect = 0.30
Identities = 15/25 (60%), Positives = 15/25 (60%)
Frame = +3
Query: 191 FFSAAMGIGNTSNAPITGQPYQSSA 215
F G NAPITGQPYQSSA
Sbjct: 624 FLQCGYGYREYFNAPITGQPYQSSA 698
>BM779563 similar to PIR|T52141|T52 hypothetical protein [imported] -
Arabidopsis thaliana, partial (41%)
Length = 564
Score = 286 bits (733), Expect = 7e-78
Identities = 146/167 (87%), Positives = 150/167 (89%)
Frame = +1
Query: 3 QNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSL 62
QNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSL
Sbjct: 1 QNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSL 180
Query: 63 FVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARE 122
FVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARE
Sbjct: 181 FVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARE 360
Query: 123 CEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPL 169
CEEVANLFSRILNAYAKVPP L+ F P++A + PL
Sbjct: 361 CEEVANLFSRILNAYAKVPPMLQ------PFPPPTPPPSLAPVSSPL 483
Score = 54.7 bits (130), Expect(2) = 8e-17
Identities = 23/23 (100%), Positives = 23/23 (100%)
Frame = +1
Query: 308 PMLQPFPPPTPPPSLAPVSSPLP 330
PMLQPFPPPTPPPSLAPVSSPLP
Sbjct: 418 PMLQPFPPPTPPPSLAPVSSPLP 486
Score = 49.7 bits (117), Expect(2) = 8e-17
Identities = 24/24 (100%), Positives = 24/24 (100%)
Frame = +3
Query: 331 NPAISREKVRDALLVLVQDNQFID 354
NPAISREKVRDALLVLVQDNQFID
Sbjct: 486 NPAISREKVRDALLVLVQDNQFID 557
>TC82579 similar to PIR|T52141|T52141 hypothetical protein [imported] -
Arabidopsis thaliana, partial (37%)
Length = 703
Score = 262 bits (670), Expect = 1e-70
Identities = 134/193 (69%), Positives = 155/193 (79%)
Frame = +2
Query: 1 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 60
MSQ KL PNLDQ STK+LNLTVLQR+DPF++EIL TAAHV+FY+FNI+ +QWSRKDVEG
Sbjct: 116 MSQTKKLTPNLDQHSTKMLNLTVLQRMDPFIDEILFTAAHVSFYDFNIETNQWSRKDVEG 295
Query: 61 SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNA 120
SLFVVKRN QPRFQFIVMNRRNT+NLVENLL DFEYE++ PYLLYRNA+QEVNGIWFYN
Sbjct: 296 SLFVVKRNAQPRFQFIVMNRRNTDNLVENLL-DFEYELKKPYLLYRNASQEVNGIWFYNP 472
Query: 121 RECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVA 180
ECEEVANLF+RILNAY K P +SS KSEFEE + V A+ + +E +SS
Sbjct: 473 EECEEVANLFNRILNAYPKALPNATMSSNKSEFEEHDQV--SAINESAIE-TSSAAGAAT 643
Query: 181 DVPDDPSFINFFS 193
D +D F NFF+
Sbjct: 644 DAVEDSVFTNFFN 682
>TC93738 similar to GP|23617133|dbj|BAC20813. transcription factor-like
protein {Oryza sativa (japonica cultivar-group)},
partial (11%)
Length = 448
Score = 164 bits (415), Expect = 5e-41
Identities = 78/78 (100%), Positives = 78/78 (100%)
Frame = +3
Query: 289 IPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLVQ 348
IPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLVQ
Sbjct: 9 IPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLVQ 188
Query: 349 DNQFIDMVYKALLNAHQS 366
DNQFIDMVYKALLNAHQS
Sbjct: 189 DNQFIDMVYKALLNAHQS 242
>TC83203
Length = 614
Score = 77.8 bits (190), Expect = 6e-15
Identities = 36/56 (64%), Positives = 45/56 (80%), Gaps = 1/56 (1%)
Frame = +2
Query: 312 PFPPPTPPPSLAPVSSPLPN-PAISREKVRDALLVLVQDNQFIDMVYKALLNAHQS 366
PFPPP PP SL P+SS +PN P ISR+KVRDAL+ LVQD++FIDM+++ LL H S
Sbjct: 2 PFPPPNPPSSLTPISSSMPNRPVISRDKVRDALVSLVQDDKFIDMMFQTLLKMHNS 169
>TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 -
Arabidopsis thaliana, partial (3%)
Length = 757
Score = 50.4 bits (119), Expect = 1e-06
Identities = 53/188 (28%), Positives = 81/188 (42%), Gaps = 3/188 (1%)
Frame = +1
Query: 147 SSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPI 206
+ST+S + +V T + + PS S+ S A P P S + S AP+
Sbjct: 4 TSTRSTSAPMASV-TSSSLARRTAPSPSLPSQSAPSPCPPR-----SPPAPVVFPSPAPV 165
Query: 207 TGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLV 266
+ + PV+P P L +P LP+ P +P + + + LV
Sbjct: 166 PAVLLSLALVLPVLLSPALVLPVLPSPVLALPVLPSPVPVPLALPSPALMPVAPLSPALV 345
Query: 267 KPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP---SLA 323
P+ PLS A ++ PV +P+ PP RP + P PP +PPP SL+
Sbjct: 346 LPA----PLSPALVL--PV---LPSPPP--------RPAALLIPDPAPPTSPPPAPVSLS 474
Query: 324 PVSSPLPN 331
P S+PLP+
Sbjct: 475 PASAPLPS 498
Score = 38.9 bits (89), Expect = 0.003
Identities = 35/139 (25%), Positives = 54/139 (38%), Gaps = 16/139 (11%)
Frame = +1
Query: 213 SSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLV-----K 267
+S T SS A +P +P + P P S P +P + + + LV
Sbjct: 34 ASVTSSSLARRTAPSPSLPSQSAPSPCPPRSPPAPVVFPSPAPVPAVLLSLALVLPVLLS 213
Query: 268 PSFFVPPLSSAAMMMPPVSSSIPT-----------APPLHPTGTVQRPYGTPMLQPFPPP 316
P+ +P L S + +P + S +P PL P + P ++ P P
Sbjct: 214 PALVLPVLPSPVLALPVLPSPVPVPLALPSPALMPVAPLSPALVLPAPLSPALVLPVLP- 390
Query: 317 TPPPSLAPVSSPLPNPAIS 335
+PPP A + P P P S
Sbjct: 391 SPPPRPAALLIPDPAPPTS 447
>TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (57%)
Length = 897
Score = 50.1 bits (118), Expect = 1e-06
Identities = 38/104 (36%), Positives = 48/104 (45%), Gaps = 4/104 (3%)
Frame = +3
Query: 236 QIPSL-PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPP 294
Q PS PT++P TP P S ++ T P+ P SS PP +S AP
Sbjct: 102 QSPSAAPTTSPSTPAATTPVSSPVAAPPTTPTTPAPVASPKSS-----PPATSPKAAAPT 266
Query: 295 LHPTGTVQRPYGTPMLQPFPPPTP---PPSLAPVSSPLPNPAIS 335
P P TP+ P P P P PP+ APVSSP PA++
Sbjct: 267 ATPPAASSPPAVTPVSTPPPAPVPVKSPPTPAPVSSP---PAVT 389
Score = 45.8 bits (107), Expect = 3e-05
Identities = 41/150 (27%), Positives = 63/150 (41%), Gaps = 2/150 (1%)
Frame = +3
Query: 187 SFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHA--ATPVVPLPTLQIPSLPTST 244
+FI A +G + S AP T P +AT S P A TP P P S P +T
Sbjct: 66 AFICIIVAGVGGQSPSAAPTTS-PSTPAATTPVSSPVAAPPTTPTTPAPVASPKSSPPAT 242
Query: 245 PFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRP 304
+P+ AP + ++ + V P PP + + PP + + + P + P V P
Sbjct: 243 --SPKAAAPTATPPAASSPPAVTP-VSTPPPAPVPVKSPPTPAPVSSPPAVTP---VAAP 404
Query: 305 YGTPMLQPFPPPTPPPSLAPVSSPLPNPAI 334
TP + P + +P P+PA+
Sbjct: 405 TTTPAVPAPAPSKGKKNKKKHGAPAPSPAL 494
Score = 32.7 bits (73), Expect = 0.23
Identities = 20/53 (37%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Frame = +3
Query: 287 SSIPTAPPLHPTGTVQRPYGTPMLQPFP-PPTPPPSLAPVSSPLPNPAISREK 338
S+ PT P P T TP+ P PPT P + APV+SP +P + K
Sbjct: 111 SAAPTTSPSTPAAT------TPVSSPVAAPPTTPTTPAPVASPKSSPPATSPK 251
>TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 - alfalfa,
partial (97%)
Length = 1049
Score = 48.9 bits (115), Expect = 3e-06
Identities = 43/144 (29%), Positives = 57/144 (38%), Gaps = 7/144 (4%)
Frame = +3
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQ 261
+N P T P S + SS P ++P P LQ P + P +P + S
Sbjct: 288 ANTPPT-TPQASPPPVQSSPPPVQSSP----PPLQSSPPPAQSTPPPVQSSPPPVQQSPP 452
Query: 262 ATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPS 321
T L P P+ S PP +S P +PP P TP PP TPPP+
Sbjct: 453 PTPLTPP-----PVQST----PPPASPPPASPPPFSPPPATPPPATP-----PPATPPPA 590
Query: 322 LA-------PVSSPLPNPAISREK 338
L P ++P P PA + K
Sbjct: 591 LTPTPLSSPPATTPAPAPAKLKSK 662
Score = 38.5 bits (88), Expect = 0.004
Identities = 33/121 (27%), Positives = 45/121 (36%), Gaps = 9/121 (7%)
Frame = +3
Query: 219 SSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSA 278
S+ P + P P P P+ P ++P P +P + SS PPL S+
Sbjct: 246 STSPNSSPAPPTP-PANTPPTTPQASP-PPVQSSPPPVQSSP------------PPLQSS 383
Query: 279 AMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQP---------FPPPTPPPSLAPVSSPL 329
PP + S P P Q P TP+ P PPP PP +P +
Sbjct: 384 ----PPPAQSTPPPVQSSPPPVQQSPPPTPLTPPPVQSTPPPASPPPASPPPFSPPPATP 551
Query: 330 P 330
P
Sbjct: 552 P 554
Score = 38.1 bits (87), Expect = 0.005
Identities = 36/129 (27%), Positives = 46/129 (34%), Gaps = 5/129 (3%)
Frame = +3
Query: 210 PYQSSATISSSGPTHAATPVVPLPTLQIP--SLPTSTPFTP---QHDAPESINSSSQATN 264
P QSS S P A + P+ + P P TP TP Q P + +
Sbjct: 348 PVQSSPPPLQSSPPPAQSTPPPVQSSPPPVQQSPPPTPLTPPPVQSTPPPASPPPASPPP 527
Query: 265 LVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAP 324
P PP + PP + P + P P TP P + P+LAP
Sbjct: 528 FSPPPATPPPATPPPATPPPALTPTPLSSP---------PATTPAPAPAKLKSKAPALAP 680
Query: 325 VSSPLPNPA 333
V SP PA
Sbjct: 681 VLSPSDAPA 707
Score = 28.5 bits (62), Expect = 4.3
Identities = 23/90 (25%), Positives = 43/90 (47%), Gaps = 11/90 (12%)
Frame = +2
Query: 186 PSFINFFSAAMGIGNTSNA--PITGQPYQSS--------ATISSSGPTHAAT-PVVPLPT 234
PS + FS+ + +TS++ P T + S + ISSS PTH++T V
Sbjct: 320 PSTCSIFSSTRPVFSTSSSVIPSTSSVHSPSRSVFSSTCSAISSSNPTHSSTCSVYSTTC 499
Query: 235 LQIPSLPTSTPFTPQHDAPESINSSSQATN 264
+ P L ++ +P + + SS+ +++
Sbjct: 500 ITTPCLTSTILSSPCYSTSRNSTSSNSSSS 589
>BG648462 weakly similar to GP|14253163|em VMP4 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 678
Score = 47.4 bits (111), Expect = 9e-06
Identities = 40/108 (37%), Positives = 49/108 (45%), Gaps = 5/108 (4%)
Frame = +1
Query: 228 PVVPLPTLQIPSLPTSTPFTPQHDAP--ESINSSSQATNL-VKPSFFVPPLSSAAMMMPP 284
PV T+ +P+ ++PF P P + + Q N + PS PPLSS +PP
Sbjct: 280 PVNQYQTVVMPNNFNASPFIPVSVTPLAQIQGTPMQPYNQQIPPSIVPPPLSS----LPP 447
Query: 285 VSSSIPTAPPLHPTGTVQRPYGTPMLQP--FPPPTPPPSLAPVSSPLP 330
P PP HP Q P P QP PPP PP S P S PLP
Sbjct: 448 PQ---PEMPPPHPHPPSQPPL--PETQPPLVPPPPPPGSPPPPSPPLP 576
>BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (23%)
Length = 1217
Score = 47.4 bits (111), Expect = 9e-06
Identities = 52/197 (26%), Positives = 72/197 (36%), Gaps = 2/197 (1%)
Frame = -3
Query: 141 PPKLKVSSTKSEFEELEAVPTMAVMDGPL--EPSSSITSNVADVPDDPSFINFFSAAMGI 198
PP L +S T PT A P+ P S S + P P ++
Sbjct: 630 PPSLDISITAPSTRPSSLTPTPASPPPPIPRSPHPSSFSPHSRSPPRPPWLR-------- 475
Query: 199 GNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINS 258
P P S++ SS P A +P P P P TP P+H A +++
Sbjct: 474 ------PFNIHPLISNSAFPSSSP--AELRFLPPPR---PRAPRPTPSFPRHSAAPPLHA 328
Query: 259 SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTP 318
+ P F+ P S A++ P+S P L P + PPPTP
Sbjct: 327 PYRL-----PHLFLHPHSPPAVI--PLSLLPVQGPDLPPVSS-------------PPPTP 208
Query: 319 PPSLAPVSSPLPNPAIS 335
PP AP P P++S
Sbjct: 207 PPPPAPPRPAAPPPSLS 157
Score = 39.7 bits (91), Expect = 0.002
Identities = 37/115 (32%), Positives = 44/115 (38%), Gaps = 6/115 (5%)
Frame = -1
Query: 226 ATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPV 285
+TP P+P P P + F P H AP + + P F+P + S P
Sbjct: 464 STPSSPIPPSH-PLHPLNCAFFP-HPAPAPRGPLPPSPAIPLPPRFMPLIGS------PT 309
Query: 286 SSSIPTAPPLHPTGTVQR---PYGTPMLQPFPPPT---PPPSLAPVSSPLPNPAI 334
SSSI PP R P P L P PPP PPP P P P I
Sbjct: 308 SSSIRIPPPRSSPYRCSRSKAPISPPFLLPPPPPRPPPPPPGRPRPPHPSPFPLI 144
Score = 37.4 bits (85), Expect = 0.009
Identities = 39/125 (31%), Positives = 48/125 (38%)
Frame = -1
Query: 205 PITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATN 264
P+T P +T S S P P PLP P L + P TP+ P A
Sbjct: 650 PLTPYPPLPLST-SLSLPHPPVRPPSPLPPPHHPLL-SPVPPTPRRSLP------IPAHP 495
Query: 265 LVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAP 324
L P F A+ SS IP + PLHP P+ P + PP+P L P
Sbjct: 494 LAHPGF--------ALSTSTPSSPIPPSHPLHPLNCAFFPHPAPAPRGPLPPSPAIPLPP 339
Query: 325 VSSPL 329
PL
Sbjct: 338 RFMPL 324
Score = 37.4 bits (85), Expect = 0.009
Identities = 58/255 (22%), Positives = 93/255 (35%), Gaps = 58/255 (22%)
Frame = -2
Query: 134 LNAYAKVPPKLKVSSTKSEFE-----ELEAVPTMAVMDGPLEPSSSITSNVADVPD---- 184
L+ + PP L++SS+ F +++ + + A +D P P + + S+++ +
Sbjct: 919 LSLSRRPPPSLRISSSPPTFSSRFAHDVDTLCSPASVDSPAHPHAPLLSDLSTIATISTP 740
Query: 185 ----DPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSG-----PTHAATPVVPLPTL 235
P + A + S++P + S S P H + P P L
Sbjct: 739 FHALSPLSLRLSPALFSPTSDSHSPPSAALPLPHIPPSLSRHLYHCPIHPSVLPHPYPRL 560
Query: 236 QIPSLP---------TSTPFTP--------QHDAPE-------------SINSSSQ---- 261
PS P + P TP QH P +++S +
Sbjct: 559 TTPSYPPFPPPLVVLSPFPLTPSPTLASPFQHPPPHLQFRLPILFTR*TALSSPTPPPRP 380
Query: 262 -ATNLVKPSFFVPPLSSAAMMMPPVS-SSIPTA--PPLHPTGTVQRPYGTPML--QPFPP 315
A +L+ P F PP S PP+ S+ P PP+ G RP P P PP
Sbjct: 379 AAHSLLPPPFRCPPASCPLSAPPPLPPSAFPPRGHPPIAAPGP--RPRSPPRFFSPPHPP 206
Query: 316 PTPPPSLAPVSSPLP 330
P P A + P+P
Sbjct: 205 APPRPPPAGRAPPIP 161
Score = 32.3 bits (72), Expect = 0.30
Identities = 37/140 (26%), Positives = 51/140 (36%), Gaps = 24/140 (17%)
Frame = -3
Query: 217 ISSSGPTHAATPVV----------PLPTLQIPSL-PTSTPFT------PQH-------DA 252
+ +S P +PVV P P++ P L P S+P + P H
Sbjct: 891 LCASPPLRLRSPVVSHTTSILYAPPPPSILPPILTPLSSPISLRSLLSPLHFTLSRLSRC 712
Query: 253 PESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQP 312
++SS L+ P +PP S + PP TAP P+ P P P
Sbjct: 711 VSHLHSSPPPPTLIHP---LPPPSPYPISPPPSLDISITAPSTRPSSLTPTPASPP--PP 547
Query: 313 FPPPTPPPSLAPVSSPLPNP 332
P P S +P S P P
Sbjct: 546 IPRSPHPSSFSPHSRSPPRP 487
Score = 31.2 bits (69), Expect = 0.66
Identities = 30/112 (26%), Positives = 50/112 (43%), Gaps = 5/112 (4%)
Frame = -3
Query: 223 THAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQ-ATNLVKPSF--FVPPLSSAA 279
T A V +L IP + ++ A I+S S ++L+ P+ PPL+
Sbjct: 1104 TSRARRVRHYTSLSIPLITVGLQYSHSAYAHCEISSRSPIVSSLLFPADRDATPPLTLRH 925
Query: 280 MMMPPVSSSIP--TAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPL 329
+ +++ +P +PPL V + + P PP PP L P+SSP+
Sbjct: 924 SLSLSLAALLPLCASPPLRLRSPVVSHTTSILYAPPPPSILPPILTPLSSPI 769
>BQ751144 weakly similar to PIR|PQ0479|PQ04 pistil extensin-like protein
(clone pMG14) - common tobacco (fragment), partial (11%)
Length = 632
Score = 47.4 bits (111), Expect = 9e-06
Identities = 37/120 (30%), Positives = 57/120 (46%), Gaps = 1/120 (0%)
Frame = +1
Query: 216 TISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPL 275
++ SS P+ + +P L+ P+ P + P +P +P +S ++ ++L P PP
Sbjct: 181 SLPSSPPSTSPSPHA---LLRSPTAPPAAPLSPPASSP---SSPTRPSSLFSP----PPP 330
Query: 276 SSAAMMMPPVSSSIPTAP-PLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAI 334
S + PP S+ P P PL P T PFPP +PPP P SSP +P +
Sbjct: 331 SPSRSRPPPPSTPSPLLPLPLLPLATEP--------SPFPPSSPPPR--PASSPSLSPPV 480
Score = 44.7 bits (104), Expect = 6e-05
Identities = 44/136 (32%), Positives = 59/136 (43%), Gaps = 13/136 (9%)
Frame = +1
Query: 213 SSATISSSGPTHAATPVVPLPTL--QIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSF 270
S + ++ P+ + PLP L +PS PT P S S+S + + + S
Sbjct: 61 SRTLVQNNRPSQPSK*STPLPFLLSPVPSWPTVVTRRRLLSLPSSPPSTSPSPHALLRSP 240
Query: 271 FVPPLSSAAMMMPPVSS--------SIPTAPPLHPTGTVQRPYGTPM-LQPFP--PPTPP 319
PP AA + PP SS S+ + PP P+ + P TP L P P P
Sbjct: 241 TAPP---AAPLSPPASSPSSPTRPSSLFSPPPPSPSRSRPPPPSTPSPLLPLPLLPLATE 411
Query: 320 PSLAPVSSPLPNPAIS 335
PS P SSP P PA S
Sbjct: 412 PSPFPPSSPPPRPASS 459
Score = 43.9 bits (102), Expect = 1e-04
Identities = 32/104 (30%), Positives = 46/104 (43%), Gaps = 3/104 (2%)
Frame = +1
Query: 210 PYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPS 269
P S + S P +P++PLP L + + P+ P + P S S S L PS
Sbjct: 319 PPPPSPSRSRPPPPSTPSPLLPLPLLPLATEPSPFPPSSPPPRPASSPSLSPPVLLFPPS 498
Query: 270 FFVPPLSSA---AMMMPPVSSSIPTAPPLHPTGTVQRPYGTPML 310
PP S + + + PP S+P +PP P + RP P L
Sbjct: 499 ASPPPWSPSRPPSALAPPSRPSLPASPPSSPRSST-RPSRFPAL 627
Score = 30.0 bits (66), Expect = 1.5
Identities = 34/132 (25%), Positives = 48/132 (35%), Gaps = 2/132 (1%)
Frame = +3
Query: 201 TSNAP-ITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSS 259
TS AP +T P +S+ SS P + +P+ T +P +TP
Sbjct: 216 TSCAPEVTNCPARSTVVSSSVVPVITNSAELPVLTTTAVPVPVTTPAA------------ 359
Query: 260 SQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHP-TGTVQRPYGTPMLQPFPPPTP 318
V P+ SA PP + PP+ P T T +P P +
Sbjct: 360 ------------VYPIPSAPAPAPPAGNGTVPVPPVKPTTETGVKPVPVTTGAAVPSVSK 503
Query: 319 PPSLAPVSSPLP 330
P +L VS P P
Sbjct: 504 PATL--VSQPAP 533
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 46.6 bits (109), Expect = 2e-05
Identities = 45/154 (29%), Positives = 63/154 (40%), Gaps = 30/154 (19%)
Frame = -3
Query: 222 PTHAATPVVPLPTLQIPSLPTST---PFTPQHDAPESINSSSQATNLVKPSFFVPPLSSA 278
PTH P +P P IPS P T P P + ++ ++ P PP +SA
Sbjct: 523 PTHQPHPCLPPP---IPSAPPPTRAFALPPSAPPPSDLPPPPRSPTVIDPR---PPAASA 362
Query: 279 AMM----MPPVSSSIPTAPP---------LHPTGTVQRPYGTPMLQ--------PFPPPT 317
+ + P+ S P PP L P+ + P P+ P PPPT
Sbjct: 361 VPLPLRPLYPLRDSPPRRPPPPPPHSPSILPPSSRLSPPTPLPLSSSP*PALPLPLPPPT 182
Query: 318 PP-PSLAPVSSPLPNPAI-----SREKVRDALLV 345
PP P AP ++P+ PA SR K D L++
Sbjct: 181 PPPPPPAPTTTPI*APAAGYTPESR*K*DDVLII 80
Score = 45.1 bits (105), Expect = 4e-05
Identities = 48/166 (28%), Positives = 66/166 (38%), Gaps = 14/166 (8%)
Frame = -1
Query: 187 SFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPS--LPTST 244
SF+ SA I + N P + QS AT++ PT + TP P P+ P P+ +
Sbjct: 606 SFLGTGSAPPAIHHHPNPPAS----QSPATLNR--PT-SHTPACPPPSRPPPHPRAPSLS 448
Query: 245 PFTPQHDAPESINSSSQATNLVKP----SFFVPPLSSAAMMMPPVS---SSIPTAPPLHP 297
P P H ++ + P F P +PP + PT PP P
Sbjct: 447 PPPPPHPPTSPPRPGRPRSSTLAPPPHRQFHSPSALCTHCGIPPPAVPPPPPPTPPPSSP 268
Query: 298 TGTVQRP-----YGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREK 338
V P + P QP P P+PPP P+ P P P SR +
Sbjct: 267 PALVSPPPRPFLFPPPPNQPSPSPSPPPP-PPLPPPPPQPPQSRHQ 133
Score = 35.4 bits (80), Expect = 0.035
Identities = 36/134 (26%), Positives = 47/134 (34%), Gaps = 19/134 (14%)
Frame = -3
Query: 218 SSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
S +G T A P Q P+ P P P + Q + P P +
Sbjct: 643 SGAGSTGHAIPYFVFGNRQRPTRHPPPP*PPSIPIPRHPEPTHQPHPCLPPPIPSAPPPT 464
Query: 278 AAMMMPPVS---SSIPTAPPLHPTGTVQRP----------------YGTPMLQPFPPPTP 318
A +PP + S +P PP PT RP +P +P PPP
Sbjct: 463 RAFALPPSAPPPSDLP-PPPRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPPPPPPH 287
Query: 319 PPSLAPVSSPLPNP 332
PS+ P SS L P
Sbjct: 286 SPSILPPSSRLSPP 245
Score = 34.7 bits (78), Expect = 0.060
Identities = 32/112 (28%), Positives = 42/112 (36%)
Frame = -1
Query: 221 GPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAM 280
GP+ A P P P + + T F AP +I+ P+ P S
Sbjct: 675 GPS-LAHPARPRPGRVLLVMQYPTSFLGTGSAPPAIHHHPNPPASQSPATLNRPTSHTPA 499
Query: 281 MMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
PP + PP HP + P +P P PP +PP P SS L P
Sbjct: 498 CPPP------SRPPPHP----RAPSLSPPPPPHPPTSPPRPGRPRSSTLAPP 373
Score = 33.5 bits (75), Expect = 0.13
Identities = 55/231 (23%), Positives = 84/231 (35%), Gaps = 29/231 (12%)
Frame = -2
Query: 131 SRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFIN 190
+R L + + PP L T L PT PL PS +I +A P P+ ++
Sbjct: 1139 TRSLPYHPRPPPPLATDHTL-----LSPRPTPRATH-PLSPSHTIP-RLAPSPGLPAPLS 981
Query: 191 FFSAAMG-----IGNTSNAPITGQPYQSSA-------TISSSGPTHAATPVVPLPTLQIP 238
+ ++ +T P P S + T+ S P A P++ P+ P
Sbjct: 980 RYYTSLSPPPLHTHDTLTPPSAPAPLPSISSRRLLRPTLHLSSPPARALPILHRPSAVNP 801
Query: 239 SLPTSTPFTPQHDAP-------ESINSSSQATNLVKPSFFVPPLSSAAMMMPPV------ 285
L TP H E++++ V P+ PP S ++PP
Sbjct: 800 PLRDPTPSLYAHALMSARIRDCETLHTPYS*RLTVHPA---PPTSDHLWLIPPARVRGGF 630
Query: 286 ----SSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
++ + P P P T L P P PPP P ++PLP P
Sbjct: 629 YWSCNTLLRFWEPAAP-----HPPSTTTLTPQHPNPPPP*TDPPATPLPAP 492
Score = 32.3 bits (72), Expect = 0.30
Identities = 24/102 (23%), Positives = 39/102 (37%)
Frame = -1
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQ 261
++ P G+P S+ H+ + + + P++P P TP +P ++ S
Sbjct: 423 TSPPRPGRPRSSTLAPPPHRQFHSPSALCTHCGIPPPAVPPPPPPTPPPSSPPALVSPPP 244
Query: 262 ATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQR 303
L P PP + PP +P PP P QR
Sbjct: 243 RPFLFPP----PPNQPSPSPSPPPPPPLPPPPPQPPQSRHQR 130
Score = 28.9 bits (63), Expect = 3.3
Identities = 35/129 (27%), Positives = 47/129 (36%), Gaps = 3/129 (2%)
Frame = -1
Query: 205 PITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATN 264
P T +P + + + P H + P P P P P+ P S+ SS
Sbjct: 1311 PSTRRPIRRLSPRVRTRPRHTSG*FRPRPP------PARAPRLPRSTHPCSVVSSLVVVR 1150
Query: 265 LVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAP 324
+ + P+SSA+ PP P P P P + P P P P P A
Sbjct: 1149 STPHT--LAPISSAS---PPSPRHRPYPPLTSPHTARYSPSLSLSHYPAPRPVPRPPRAA 985
Query: 325 VS---SPLP 330
VS PLP
Sbjct: 984 VSILHLPLP 958
>TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (15%)
Length = 984
Score = 46.6 bits (109), Expect = 2e-05
Identities = 42/133 (31%), Positives = 59/133 (43%), Gaps = 5/133 (3%)
Frame = +1
Query: 209 QPYQSSATISSSGPTHAATPVVPLPTLQIPS-LPTSTPFTPQHDAPESINSSSQATNLVK 267
QP S + + P + +P+ PL + PS LP +TP P S +S + +
Sbjct: 217 QP*LPSPLMEAPSPRPSPSPLSPL---RFPSRLPLTTP--PSLTTSPSPTASPPPRSPPR 381
Query: 268 PSFFVPPLSSAAMMMPPVSS--SIPTAPPLHPTGTVQR-PYGTPMLQPFPPPTPPPSLAP 324
P PP S +M P P+ PP +P + P P + P P PT PP+ P
Sbjct: 382 P----PPRSPPRQLMSPPRPLPRSPSPPPRNPPELSRTTPTPRPPMSPSPSPTSPPAATP 549
Query: 325 -VSSPLPNPAISR 336
VS P P PA+ R
Sbjct: 550 PVSRPPPRPALRR 588
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 46.2 bits (108), Expect = 2e-05
Identities = 35/112 (31%), Positives = 52/112 (46%), Gaps = 4/112 (3%)
Frame = +3
Query: 221 GPTHAATPVVPLPTLQIPSL-PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAA 279
G + ++ P PT+ PS P + P P+ AP + SS + + P +S AA
Sbjct: 117 GQSPSSAPTTSPPTVTTPSAAPVAAPTKPKSPAPVASPKSSPPASSPTAATVTPAVSPAA 296
Query: 280 MMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTP---PPSLAPVSSP 328
+ P S ++P + P T +P P+ P P P P PP+ PVSSP
Sbjct: 297 PV-PVAKSPAASSPVVAPVSTPPKP--APVSSP-PAPVPVSSPPTPVPVSSP 440
Score = 32.0 bits (71), Expect = 0.39
Identities = 16/54 (29%), Positives = 24/54 (43%)
Frame = +3
Query: 275 LSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSP 328
L+ ++ V P++ P TV P P+ P P +P P +P SSP
Sbjct: 81 LALICIVFAGVGGQSPSSAPTTSPPTVTTPSAAPVAAPTKPKSPAPVASPKSSP 242
>TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk protein
{Argiope trifasciata}, partial (3%)
Length = 1177
Score = 46.2 bits (108), Expect = 2e-05
Identities = 45/162 (27%), Positives = 58/162 (35%), Gaps = 31/162 (19%)
Frame = +3
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFT------------- 247
T+ P T Y S + S+ PT ++P P S P+++P
Sbjct: 3 TTRTPSTSPSYASRTPLRSTPPTRPSSPTPSAPRTSSRSTPSTSPSRRPRSKTPLPPPPP 182
Query: 248 --PQHDAP----------ESINSSSQATNLVKPSFFVPPLSSAAMM----MPPVSSSIPT 291
PQH P + SS A + P PP S + P S+ PT
Sbjct: 183 PLPQHPKPPQKSPPRMRRSKPSPSSPAATPLWPQRTTPPPSPSTPRPSP*TPATPSTSPT 362
Query: 292 AP-PLHPTGTVQRPYGTPM-LQPFPPPTPPPSLAPVSSPLPN 331
AP P P T P TP P PPTP P VS P+
Sbjct: 363 APLPTPPPRTTLPPALTPRPPSPSTPPTPKPGPVSVSPASPS 488
>TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrate lyase
subunit 2 {Neurospora crassa}, complete
Length = 2389
Score = 45.8 bits (107), Expect = 3e-05
Identities = 42/131 (32%), Positives = 58/131 (44%), Gaps = 11/131 (8%)
Frame = +2
Query: 209 QPYQSSATISSSGPTHAATPV----------VPLPTLQIPSLPTSTPFTPQHDAPESINS 258
QP + SSS P A+ + P+P + S+P Q+ A +
Sbjct: 368 QPL*LAPARSSSNPAPGASDLFFRVQCNHKYAPIPK---STADESSPKPDQNKAGKRTPK 538
Query: 259 SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPP-PT 317
S+ NL+ +FF+PP PP SS PTA P T + P +P+L P PP T
Sbjct: 539 KSR--NLL--TFFIPP------ECPPSPSSRPTARPSSTTTSPVLPSSSPLLSPLPPSTT 688
Query: 318 PPPSLAPVSSP 328
PPP A +SP
Sbjct: 689 PPPDCALCTSP 721
Score = 40.4 bits (93), Expect = 0.001
Identities = 40/134 (29%), Positives = 55/134 (40%), Gaps = 25/134 (18%)
Frame = +3
Query: 218 SSSGPTHAATPVVPLPTLQIPS------LPTSTPFTPQH----DAPESINSSSQATNLVK 267
S+ PT +A PLP L +PS LPT+ T +P S L
Sbjct: 1320 STRPPTLSAVSSGPLPALPLPSA*PTLPLPTAKRSTSMRARRWSSPLLSVVS*PRRRLTS 1499
Query: 268 PSFF---VPPLS------------SAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQP 312
P+ VPP S S+ +++PP S+ P+ PP PT + T +L P
Sbjct: 1500 PTLMPRPVPPSSLLSSTPTAVSGLSSPVVVPPSSTPTPSPPPASPTSS-PTTVSTLVLPP 1676
Query: 313 FPPPTPPPSLAPVS 326
P PT P L+ S
Sbjct: 1677 SPRPTTTPVLSSTS 1718
>TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean, partial
(93%)
Length = 1554
Score = 45.1 bits (105), Expect = 4e-05
Identities = 42/132 (31%), Positives = 58/132 (43%), Gaps = 6/132 (4%)
Frame = -2
Query: 212 QSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHD---APESINSSSQATNLVKP 268
Q+ AT + P T + P+ T Q P++ S P T Q AP+S +S A +
Sbjct: 854 QAPATAPTKLPPTTPTAITPVTT-QPPTVVASPPITSQPPVTVAPKSAPVTSPAPKIAPA 678
Query: 269 SF-FVPPLSSAAMMMPPVSS--SIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPV 325
S VPP PP SS S PT PP P P + P P TPP
Sbjct: 677 SSPKVPP------PQPPKSSPVSTPTLPPPLPP--------PPKISPTPVQTPPAPAPVK 540
Query: 326 SSPLPNPAISRE 337
++P+P PA +++
Sbjct: 539 ATPVPAPAPAKQ 504
Score = 34.3 bits (77), Expect = 0.078
Identities = 42/162 (25%), Positives = 57/162 (34%), Gaps = 27/162 (16%)
Frame = -2
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIP-SLPTSTP-FTPQHDAPESINS 258
TS P+T P + T + A++P VP P Q P S P STP P P I+
Sbjct: 752 TSQPPVTVAPKSAPVTSPAPKIAPASSPKVPPP--QPPKSSPVSTPTLPPPLPPPPKISP 579
Query: 259 SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLH-PTGTVQRPYGTPMLQ------ 311
+ T P+ + A + + T+PP+ PT ++ P P
Sbjct: 578 TPVQTPPAPAPVKATPVPAPAPAKQAPTPAPATSPPIPAPTPAIEAPVPAPESSKPKRRR 399
Query: 312 ------------PFP------PPTPPPSLAPVSSPLPNPAIS 335
P P PP PP S P PA S
Sbjct: 398 HRPKHRRHQAPAPAPTVIHKSPPAPPTDTTADSDTAPAPAPS 273
>TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2G14_40
{Arabidopsis thaliana}, partial (11%)
Length = 1320
Score = 45.1 bits (105), Expect = 4e-05
Identities = 42/133 (31%), Positives = 51/133 (37%), Gaps = 19/133 (14%)
Frame = +2
Query: 215 ATISSSGPTHAATPVVPLPTLQ-----IPSLPTST-----PFTPQHDAPES--INSSSQA 262
AT +S P P V PT+ +P + T T P P+ P S +
Sbjct: 104 ATTPTSAPVKPPVPKVATPTVAPAKPLVPKVTTPTAAPVKPIVPKTTTPTSAPLKPPVPK 283
Query: 263 TNLVKPSFFVPPL-SSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQP------FPP 315
KPS PP+ S PV S +P P L P + P TP L P P
Sbjct: 284 APAPKPSPVKPPVPKSKPPTTAPVKSPVPDPPKLAPV-KLPVPKVTPALSPKTPSPKIQP 460
Query: 316 PTPPPSLAPVSSP 328
P PP APVS P
Sbjct: 461 PPHPPKKAPVSLP 499
Score = 42.0 bits (97), Expect = 4e-04
Identities = 34/116 (29%), Positives = 49/116 (41%), Gaps = 3/116 (2%)
Frame = +2
Query: 220 SGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAA 279
S P H P+P + PTS P P P+ + + P P +
Sbjct: 59 SRPQHLLPWKSPVPKA---TTPTSAPVKPP--VPKVATPTVAPAKPLVPKVTTPTAAPVK 223
Query: 280 MMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTP---PPSLAPVSSPLPNP 332
++P ++ PT+ PL P + P P P PP P PP+ APV SP+P+P
Sbjct: 224 PIVPKTTT--PTSAPLKPP-VPKAP--APKPSPVKPPVPKSKPPTTAPVKSPVPDP 376
Score = 38.1 bits (87), Expect = 0.005
Identities = 24/68 (35%), Positives = 32/68 (46%), Gaps = 1/68 (1%)
Frame = +3
Query: 268 PSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQ-PFPPPTPPPSLAPVS 326
P+ P +S A +P + AP PT + P P + P P PT AP S
Sbjct: 546 PASKPPKVSPAPAKVPKAPAPAKEAPAPAPTHKKKAPAPAPDKETPAPAPTHKKKKAPKS 725
Query: 327 SPLPNPAI 334
SP+P+PAI
Sbjct: 726 SPVPSPAI 749
Score = 28.9 bits (63), Expect = 3.3
Identities = 35/116 (30%), Positives = 43/116 (36%), Gaps = 6/116 (5%)
Frame = +3
Query: 212 QSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPF-TPQHDAPESINSSSQATNLVKPSF 270
+ A SS P+ A P P PT I + P+S P +P+ DAPE K S
Sbjct: 705 KKKAPKSSPVPSPAILPPSPAPTPAIDT-PSSAPAPSPEDDAPEPPPPHKHKRRKHKHSK 881
Query: 271 FVPPLSSAAMMMPPVSSSI-----PTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPS 321
+ A P SSS P AP T+ G P P P PS
Sbjct: 882 HKKHHALALAPEPTSSSSTIIRRSPPAPLADDNTTMSSDEG-------PSPAPSPS 1028
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 44.7 bits (104), Expect = 6e-05
Identities = 44/151 (29%), Positives = 53/151 (34%), Gaps = 2/151 (1%)
Frame = +3
Query: 171 PSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVV 230
PSSS+ VA P S I PYQ + S S A++PV
Sbjct: 3 PSSSLPPGVAQTP------------------SGNGIAPSPYQFNNPTSPSSSPSASSPV- 125
Query: 231 PLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIP 290
T S P S+P + P S +SS P P S PVS++ P
Sbjct: 126 STTTSPPASTPASSPVSTTTSPPASTPASSPVPTTTSPPAPTPASS-------PVSTNSP 284
Query: 291 TAPPLH--PTGTVQRPYGTPMLQPFPPPTPP 319
TA P P P T P P TPP
Sbjct: 285 TASPAGSLPAAATPSPSSTTAGSPSPSSTPP 377
Score = 38.1 bits (87), Expect = 0.005
Identities = 32/100 (32%), Positives = 42/100 (42%)
Frame = +3
Query: 230 VPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSI 289
+P Q PS P Q + P S +SS A++ V + PP S+ P SS +
Sbjct: 15 LPPGVAQTPSGNGIAPSPYQFNNPTSPSSSPSASSPVSTTTS-PPAST------PASSPV 173
Query: 290 PTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPL 329
T T P TP P P T PP+ P SSP+
Sbjct: 174 ST--------TTSPPASTPASSPVPTTTSPPAPTPASSPV 269
Score = 34.7 bits (78), Expect = 0.060
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 11/74 (14%)
Frame = +3
Query: 268 PSFFVPP-----LSSAAMMMPPVSSSIPTAPPLHPTG------TVQRPYGTPMLQPFPPP 316
PS +PP S + P + PT+P P+ T P TP P
Sbjct: 3 PSSSLPPGVAQTPSGNGIAPSPYQFNNPTSPSSSPSASSPVSTTTSPPASTPASSPVSTT 182
Query: 317 TPPPSLAPVSSPLP 330
T PP+ P SSP+P
Sbjct: 183 TSPPASTPASSPVP 224
>AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (10%)
Length = 478
Score = 44.3 bits (103), Expect = 8e-05
Identities = 37/109 (33%), Positives = 47/109 (42%), Gaps = 6/109 (5%)
Frame = +1
Query: 222 PTHAATPVVPLPTLQIPSLPTSTPFTPQHDAP-ESINSSSQATNLVKPSFFVPPLSSAAM 280
P+H P+ P P+ P P +PF P + P SI S A PS PP
Sbjct: 76 PSH---PLNPPPSPFHPINPPPSPFHPFNPPPPHSIKSPPPA-----PSHSSPPPPPPPR 231
Query: 281 MMPPVSSSIPTAPPL---HPTGTVQRPYGT--PMLQPFPPPTPPPSLAP 324
PP S P +PP HP + P P ++P PPP PPS +P
Sbjct: 232 KXPPPPPSHPFSPPPPHNHPPPSPHHPIAPSPPHVRPSPPPPLPPSPSP 378
Score = 33.9 bits (76), Expect = 0.10
Identities = 24/65 (36%), Positives = 28/65 (42%), Gaps = 5/65 (7%)
Frame = +3
Query: 268 PSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPP-----PTPPPSL 322
PS PP S+A+ PP SS T PP P+ T T F P T PS
Sbjct: 66 PSSTFPPPQSSAITFPPHQSSTITFPPF*PSTTSFHQISTTSTISFIPSSSSSSTQTPST 245
Query: 323 APVSS 327
A +SS
Sbjct: 246 ATISS 260
Score = 30.8 bits (68), Expect = 0.87
Identities = 27/81 (33%), Positives = 34/81 (41%), Gaps = 7/81 (8%)
Frame = +1
Query: 259 SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTP--MLQPFPPP 316
S ATN +P+F+ P +PP P + PL+P + P P PF PP
Sbjct: 16 SKMATN--QPNFYFP-----FPHLPP-----PPSHPLNPPPSPFHPINPPPSPFHPFNPP 159
Query: 317 TP-----PPSLAPVSSPLPNP 332
P PP SSP P P
Sbjct: 160 PPHSIKSPPPAPSHSSPPPPP 222
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,182,297
Number of Sequences: 36976
Number of extensions: 214107
Number of successful extensions: 7579
Number of sequences better than 10.0: 647
Number of HSP's better than 10.0 without gapping: 3047
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4939
length of query: 366
length of database: 9,014,727
effective HSP length: 97
effective length of query: 269
effective length of database: 5,428,055
effective search space: 1460146795
effective search space used: 1460146795
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146332.11


