

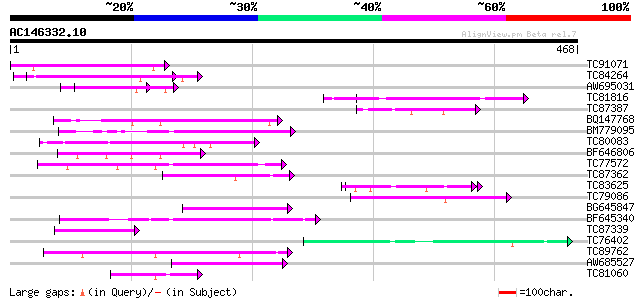
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.10 - phase: 0
(468 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91071 similar to GP|21536755|gb|AAM61087.1 unknown {Arabidopsi... 67 1e-11
TC84264 similar to PIR|G96811|G96811 unknown protein T11I11.17 [... 62 4e-10
AW695031 similar to GP|21536755|gb unknown {Arabidopsis thaliana... 56 3e-08
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 55 4e-08
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 54 2e-07
BQ147768 weakly similar to PIR|H84632|H84 probable receptor-like... 49 3e-06
BM779095 weakly similar to PIR|B84431|B8 probable receptor prote... 48 9e-06
TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 47 2e-05
BF646806 weakly similar to PIR|T02565|T025 disease resistance pr... 47 2e-05
TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C1188... 47 2e-05
TC87362 weakly similar to GP|21689861|gb|AAM67491.1 unknown prot... 47 2e-05
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 46 3e-05
TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein l... 45 6e-05
BG645847 similar to GP|11994124|d receptor protein kinase {Arabi... 44 1e-04
BF645340 weakly similar to GP|6016693|gb|A putative disease resi... 44 1e-04
TC87339 similar to PIR|D85041|D85041 hypothetical protein AT4g03... 44 1e-04
TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F2... 44 2e-04
TC89762 weakly similar to GP|7268809|emb|CAB79014.1 leucine rich... 43 3e-04
AW685527 similar to GP|1946300|emb hypothetical protein {Silene ... 42 4e-04
TC81060 similar to GP|22136192|gb|AAM91174.1 unknown protein {Ar... 41 9e-04
>TC91071 similar to GP|21536755|gb|AAM61087.1 unknown {Arabidopsis
thaliana}, partial (58%)
Length = 684
Score = 67.0 bits (162), Expect = 1e-11
Identities = 46/142 (32%), Positives = 77/142 (53%), Gaps = 10/142 (7%)
Frame = +3
Query: 1 MTRLSSEQVLKDNNAVNP-------SSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNL 53
+ LS Q L + AV P SS+ L L L ++ ++ F KL D+ FN +
Sbjct: 228 LKNLSLRQNLITDAAVVPLSSWNTLSSLEELVLRDNQLKNIPDVSIFKKLLVFDVSFNEI 407
Query: 54 TSLEGL-RACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRAL 112
SL G+ R C TLK L V +N++ +E I+ +L +L G NKL+ M+ + +L ++ L
Sbjct: 408 ASLHGVSRVCNTLKELYVSKNEVTKIEEIEHFHQLQILELGSNKLRVMENLQTLVNLQEL 587
Query: 113 ILNDN--EIVSICNLDQMKELN 132
L N ++V++C L +K+++
Sbjct: 588 WLGRNRIKVVNLCGLKCIKKIS 653
>TC84264 similar to PIR|G96811|G96811 unknown protein T11I11.17 [imported] -
Arabidopsis thaliana, partial (31%)
Length = 752
Score = 62.4 bits (150), Expect = 4e-10
Identities = 40/140 (28%), Positives = 76/140 (53%), Gaps = 5/140 (3%)
Frame = +1
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSL-EGLRAC 62
LS+ ++ + P S+ +L+L+ +S + L +L LDL +N ++ + +GL +C
Sbjct: 310 LSNNFIVTISPGCLPKSVQTLNLSRNKISTIEGLKELTRLRVLDLSYNCISRIGQGLSSC 489
Query: 63 VTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSL----STIRALILNDNE 118
+K L + +NK+ +EG+ L KLTVL+ NK+ + +G L ++++AL L N
Sbjct: 490 TIVKELYLADNKISDVEGLHRLFKLTVLDLSFNKITTTKALGQLVANYNSLQALNLLGNA 669
Query: 119 IVSICNLDQMKELNTLVLSK 138
I +Q+ + + +L K
Sbjct: 670 IQRNIGDEQLNKAVSGLLPK 729
Score = 55.5 bits (132), Expect = 4e-08
Identities = 37/149 (24%), Positives = 82/149 (54%), Gaps = 4/149 (2%)
Frame = +1
Query: 15 AVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENK 74
++NP+S S H++ + + ++ F+ L ++L N + ++ +++ L++ NK
Sbjct: 214 SLNPAS-SVAHISGVGIKAIPVISHFSNLRSVNLSNNFIVTISPGCLPKSVQTLNLSRNK 390
Query: 75 LESLEGIQGLTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIVSICNLDQMKELNT 133
+ ++EG++ LT+L VL+ N + + + + S + ++ L L DN+I + L ++ +L
Sbjct: 391 ISTIEGLKELTRLRVLDLSYNCISRIGQGLSSCTIVKELYLADNKISDVEGLHRLFKLTV 570
Query: 134 LVLSKNPI---RKIGEALKKVKSITKLSL 159
L LS N I + +G+ + S+ L+L
Sbjct: 571 LDLSFNKITTTKALGQLVANYNSLQALNL 657
>AW695031 similar to GP|21536755|gb unknown {Arabidopsis thaliana}, partial
(35%)
Length = 656
Score = 55.8 bits (133), Expect = 3e-08
Identities = 31/91 (34%), Positives = 50/91 (54%), Gaps = 5/91 (5%)
Frame = -1
Query: 54 TSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALI 113
TS+ G C+ L+ L + N + +EG+ L L VL+ NKL S+D+I +L+ + L
Sbjct: 620 TSMIGFEGCIALEELYLSHNGITKMEGLSSLANLRVLDVSSNKLTSVDDIHNLTQLEDLW 441
Query: 114 LNDNEIVSICNLDQ-----MKELNTLVLSKN 139
LNDN+I S+ + ++L T+ L N
Sbjct: 440 LNDNQIESLEGFAEAVAGSREKLTTIYLENN 348
Score = 44.7 bits (104), Expect = 8e-05
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 5/80 (6%)
Frame = -1
Query: 43 LEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDE 102
LE+L L N +T +EGL + L+ L V NKL S++ I LT+L L N+++S++
Sbjct: 587 LEELYLSHNGITKMEGLSSLANLRVLDVSSNKLTSVDDIHNLTQLEDLWLNDNQIESLEG 408
Query: 103 I-----GSLSTIRALILNDN 117
GS + + L +N
Sbjct: 407 FAEAVAGSREKLTTIYLENN 348
Score = 40.8 bits (94), Expect = 0.001
Identities = 20/61 (32%), Positives = 37/61 (59%)
Frame = -1
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
++ L+L+H ++ + L+S L LD+ N LTS++ + L+ L + +N++ESLE
Sbjct: 590 ALEELYLSHNGITKMEGLSSLANLRVLDVSSNKLTSVDDIHNLTQLEDLWLNDNQIESLE 411
Query: 80 G 80
G
Sbjct: 410 G 408
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 55.5 bits (132), Expect = 4e-08
Identities = 48/170 (28%), Positives = 77/170 (45%), Gaps = 1/170 (0%)
Frame = +3
Query: 260 IDKDTKNEKGHMTDDAHDFSFDHVDQNEDDHLEAADKRKSNKKRKETADASEKEAGVYDK 319
I+KD + K DD D +++ D E DK+ KK+KE +KE V +
Sbjct: 138 IEKDLEI-KSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKE-----KKEENVKGE 299
Query: 320 ENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKK- 378
E G K + KKK + D +KK KKD K +K +E + ++KKK
Sbjct: 300 EEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKN 479
Query: 379 KNRKNKEQSEFDIIDDAEASFAEIFNIKDQENLNHGGEMKLQDQVPKDLK 428
K++K K++ E D+ E + +I +E G E K + + ++ K
Sbjct: 480 KDKKEKKKEE----DEKEEGKVSVRDIDIEETAKEGKEKKKKKEDKEEKK 617
Score = 44.3 bits (103), Expect = 1e-04
Identities = 41/143 (28%), Positives = 65/143 (44%), Gaps = 1/143 (0%)
Frame = +3
Query: 287 EDDHLEAADKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNK-KKDKLTGTVDPDTK 345
E+ ++ KS KE EK+ + E K+ +K KKDK D K
Sbjct: 69 EESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKK 248
Query: 346 NKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEASFAEIFNI 405
+K KKK KK++N E+ +E ++ EKKKK +K K + + D + + S +
Sbjct: 249 DKEKKKKEKKEENVKGEEEDG-DEKKDK-EKKKKEKKEKGKEDKDKDGEEKKSKKDKEKK 422
Query: 406 KDQENLNHGGEMKLQDQVPKDLK 428
KD+ + GE + + KD K
Sbjct: 423 KDKNEDDDEGEDGSKKKKNKDKK 491
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 53.5 bits (127), Expect = 2e-07
Identities = 44/118 (37%), Positives = 54/118 (45%), Gaps = 16/118 (13%)
Frame = +3
Query: 287 EDDHLEAADKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGN------------KKKD 334
EDD E K+K KK KE A+ E V +KE +K+ G KKK
Sbjct: 375 EDDKKE---KKKKKKKDKENGAAASDEEKV-EKEKKKKHKEKGEDGSPEVEKSDKKKKKH 542
Query: 335 KLTGTVDPDTKNKSTKKKLKKD----DNKPSEKALALEENVNRTEKKKKNRKNKEQSE 388
K T V +KS KKK KKD DN E N +R+EKK K +KNK+ E
Sbjct: 543 KETSEVGSPEVDKSEKKKKKKDKEAKDNAADISNGNDESNADRSEKKHKKKKNKDAQE 716
>BQ147768 weakly similar to PIR|H84632|H84 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana, partial (12%)
Length = 712
Score = 49.3 bits (116), Expect = 3e-06
Identities = 56/198 (28%), Positives = 90/198 (45%), Gaps = 9/198 (4%)
Frame = +3
Query: 37 LASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNK 96
L + +KL LD +N SLEG E + +L L+ N
Sbjct: 6 LGNLSKLTHLDFSYN---SLEG-----------------EIPNSLGNHRQLKYLDISNNN 125
Query: 97 LKSM--DEIGSLSTIRALILNDNEIVSIC--NLDQMKELNTLVLSKNP-IRKIGEALKKV 151
L E+G + + +L L+ N I +L + +L LV+ N + KI ++ +
Sbjct: 126 LNGSIPHELGFIKYLGSLNLSTNRISGDIPPSLGNLVKLTHLVIYGNSLVGKIPPSIGNL 305
Query: 152 KSITKLSLSHCQLEG-IDTSLKFCVELTELRLAHNDIK-SLPEELMHNSKLRNLDLGNNV 209
+S+ L +S ++G I L LT LRL+HN IK +P L + +L LD+ NN
Sbjct: 306 RSLESLEISDNYIQGSIPPRLGLLKNLTTLRLSHNRIKGEIPPSLGNLKQLEELDISNNN 485
Query: 210 IAKW--SDIKVLKSLTKL 225
I + ++ +LK+LT L
Sbjct: 486 IQGFLPFELGLLKNLTTL 539
>BM779095 weakly similar to PIR|B84431|B8 probable receptor protein kinase
[imported] - Arabidopsis thaliana, partial (15%)
Length = 721
Score = 47.8 bits (112), Expect = 9e-06
Identities = 57/200 (28%), Positives = 93/200 (46%), Gaps = 4/200 (2%)
Frame = +3
Query: 41 NKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSM 100
+KL+ LDL FNNLT S+ + K++ + L +L + +G N
Sbjct: 78 DKLQSLDLSFNNLTG-------------SISDIKID----CKSLLQLDL--SGNN----- 185
Query: 101 DEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIR-KIGEALKKVKSITKLSL 159
L+D+ +S+ N +K LN L+ N I I +AL ++ + L L
Sbjct: 186 -------------LSDSFPISLSNCTSLKSLN---LASNFISGGIPKALGQLNKLQSLDL 317
Query: 160 SHCQLEG-IDTSL-KFCVELTELRLAHNDIK-SLPEELMHNSKLRNLDLGNNVIAKWSDI 216
SH Q+ G I + L C L EL+L+ N+I S+P + L+ +D+ NN +
Sbjct: 318 SHNQITGWIPSELSNVCGSLLELKLSFNNITGSVPFGFNSCTWLQLVDISNNNMTGELPE 497
Query: 217 KVLKSLTKLRNLNLQGNPVA 236
V++SL L+ L L N ++
Sbjct: 498 SVIRSLGSLQELRLGNNAIS 557
Score = 31.6 bits (70), Expect = 0.68
Identities = 36/137 (26%), Positives = 58/137 (42%), Gaps = 12/137 (8%)
Frame = +3
Query: 17 NPSSISSLHLTHKALSD--VSCLASFNKLEKLDLKFNNLTS---LEGLRACVTLKWLSVV 71
N +S+ SL+L +S L NKL+ LDL N +T E C +L L +
Sbjct: 216 NCTSLKSLNLASNFISGGIPKALGQLNKLQSLDLSHNQITGWIPSELSNVCGSLLELKLS 395
Query: 72 ENKLESLE--GIQGLTKLTVLNAGKNKLKS---MDEIGSLSTIRALILNDNEIVS--ICN 124
N + G T L +++ N + I SL +++ L L +N I +
Sbjct: 396 FNNITGSVPFGFNSCTWLQLVDISNNNMTGELPESVIRSLGSLQELRLGNNAISMKFPSS 575
Query: 125 LDQMKELNTLVLSKNPI 141
+ +K+L + S N I
Sbjct: 576 ISSLKKLRIVDFSSNKI 626
>TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (4%)
Length = 1269
Score = 47.0 bits (110), Expect = 2e-05
Identities = 55/192 (28%), Positives = 94/192 (48%), Gaps = 10/192 (5%)
Frame = +3
Query: 25 HLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVE-NKLESLEGIQG 83
+LTH + DVS L++ KL + +N +T + L+ LS NK+E + G
Sbjct: 279 YLTH--IPDVSGLSNLEKLS-FERCYNLITIHNSIGHLNKLERLSAYGCNKVEHFPPL-G 446
Query: 84 LTKLTVLNAGK-NKLKSMDEI-GSLSTIRALILNDNEIVSI-CNLDQMKELNTLVLSKNP 140
L L LN LKS E+ ++ I + L + I + + + EL+ L ++
Sbjct: 447 LASLKELNLSSCRSLKSFPELLCKMTNIDNIWLCNTSIGELPFSFQNLSELHKLSVTYGM 626
Query: 141 IR--KIGEALKKV--KSITKLSLSHCQL--EGIDTSLKFCVELTELRLAHNDIKSLPEEL 194
+R K + + + ++T+L+L HC L E + LK+CV +T L L +++ K LPE L
Sbjct: 627 LRFPKQNDKMYSIVFSNVTQLTLYHCNLSDECLRRVLKWCVNMTSLNLLYSNFKILPECL 806
Query: 195 MHNSKLRNLDLG 206
L +++G
Sbjct: 807 SECHHLVEINVG 842
>BF646806 weakly similar to PIR|T02565|T025 disease resistance protein
homolog At2g32660 [imported] - Arabidopsis thaliana,
partial (5%)
Length = 624
Score = 47.0 bits (110), Expect = 2e-05
Identities = 39/135 (28%), Positives = 67/135 (48%), Gaps = 13/135 (9%)
Frame = +3
Query: 40 FNKLEKLDLKFNNLT---SLEGLRACVTLKWLSVVENKLESL---EGIQGLTKLTVLNAG 93
F++L+ LDL +NNL + GL +LK L N+L L EG + L++L +LN
Sbjct: 99 FHRLKSLDLSYNNLNGSLDISGLSNLTSLKILDFTSNQLVDLIVREGSKNLSRLDILNLD 278
Query: 94 KNKLKS---MDEIGSLSTIRALILNDNEIVSIC---NLDQMKELNTLVLSKNP-IRKIGE 146
N + + + +IR L L +N+ + ++K+L L LS N + K+
Sbjct: 279 SNMINGSNLQQWLWAFPSIRNLTLRNNQFKGTILDGDWSKLKKLEELDLSGNEFVGKLPS 458
Query: 147 ALKKVKSITKLSLSH 161
+ + S+ L+LS+
Sbjct: 459 SFFNMTSLLTLNLSN 503
>TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C11888)
C26225(C11888) correspond to a region of the predicted
gene.~Similar to Oryza, partial (5%)
Length = 2535
Score = 47.0 bits (110), Expect = 2e-05
Identities = 56/220 (25%), Positives = 103/220 (46%), Gaps = 15/220 (6%)
Frame = +3
Query: 24 LHLTHKALSDVSCLASFNKLEKL-----DLKFNNLTSLEGLRACVTLKWLSVVENKLESL 78
L+L K S +A +KL L D + L ++E L + L+ + + + S
Sbjct: 1701 LNLHTKQYSLPEWIAKMSKLRVLIITNYDFHPSKLNNIELLGSLQNLERIRLERIYVPSF 1880
Query: 79 EGIQGLTKL------TVLNAGKNKLKSMDEIGSLSTIRALILNDNEI--VSICNLDQMKE 130
++ L KL T+L K + D +L + D + + IC++ +K+
Sbjct: 1881 GTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLPIGICDIFLLKK 2060
Query: 131 LNTLVLSKNPIRKIGEALKKVKSITKLSLSHC-QLEGIDTSLKFCVELTELRLAHN-DIK 188
L V + + + + + + K++++ LSLS C LE I TS+ + L L +++ +
Sbjct: 2061 LR--VTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLS 2234
Query: 189 SLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNL 228
SLPEE + L+NLD+ A + I++ S+ L+NL
Sbjct: 2235 SLPEEFGNLCNLKNLDM-----ASCASIELPFSVVNLQNL 2339
>TC87362 weakly similar to GP|21689861|gb|AAM67491.1 unknown protein
{Arabidopsis thaliana}, partial (46%)
Length = 1996
Score = 46.6 bits (109), Expect = 2e-05
Identities = 32/111 (28%), Positives = 53/111 (46%), Gaps = 2/111 (1%)
Frame = +1
Query: 127 QMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHN- 185
++ L L++ N +R + ++ ++KS+ L +L G+ + L L L+ N
Sbjct: 1156 ELPNLKKLMIQLNKMRSVPSSICELKSLCYLDAHVNELHGLPAAFGRLTTLEILNLSSNF 1335
Query: 186 -DIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPV 235
D+K LPE + L+ LD+ NN I D L L LNL+ NP+
Sbjct: 1336 ADLKELPETFGELTNLKELDVSNNQIHALPD--TFGCLDNLTKLNLEQNPL 1482
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 45.8 bits (107), Expect = 3e-05
Identities = 34/115 (29%), Positives = 58/115 (49%), Gaps = 4/115 (3%)
Frame = +2
Query: 275 AHDFSFDHVDQNEDDHLEAADK--RKSNKKRKETADASEKEAGVYDKENTGHNKDNGNKK 332
A D D V+ +D +++ +K K KK KE D ++ + G K+ K KK
Sbjct: 500 AGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEG---KDKKDKEKKKKEKK 670
Query: 333 KDKLTGTVDP--DTKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKE 385
++ + G + + K+K KKK KK+ K +K EE ++ +K+KK KN++
Sbjct: 671 EENVKGEEEDGDEKKDKEKKKKEKKEKGK-EDKDKDGEEKKSKKDKEKKKDKNED 832
Score = 45.8 bits (107), Expect = 3e-05
Identities = 35/119 (29%), Positives = 54/119 (44%), Gaps = 6/119 (5%)
Frame = +2
Query: 278 FSFDHVD-----QNEDDHLEAADKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNK- 331
F++ +VD + E+ ++ KS KE EK+ + E K+ +K
Sbjct: 419 FTYIYVDITVSSKMEESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKE 598
Query: 332 KKDKLTGTVDPDTKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFD 390
KKDK D K+K KKK KK++N E+ E+ +KK+K K KE + D
Sbjct: 599 KKDKTDVDEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKD 775
Score = 40.8 bits (94), Expect = 0.001
Identities = 32/104 (30%), Positives = 45/104 (42%)
Frame = +2
Query: 260 IDKDTKNEKGHMTDDAHDFSFDHVDQNEDDHLEAADKRKSNKKRKETADASEKEAGVYDK 319
I+KD + K DD D +++ D E DK+ KK+KE +KE V +
Sbjct: 530 IEKDLEI-KSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKE-----KKEENVKGE 691
Query: 320 ENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKPSEK 363
E G K + KKK + D +KK KKD K +K
Sbjct: 692 EEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDK 823
>TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein ligase 1
putative {Plasmodium falciparum 3D7}, partial (0%)
Length = 1418
Score = 45.1 bits (105), Expect = 6e-05
Identities = 30/144 (20%), Positives = 61/144 (41%), Gaps = 11/144 (7%)
Frame = +2
Query: 282 HVDQNEDDHLEAADKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVD 341
H + +E+ EA + ++ +E+++ + K N +N +++ T +
Sbjct: 116 HDENDEETSSEAGSSSEGGEEEEESSEEEAPKTTPPPKTNPKNNSVEDEDEEETDTDEDE 295
Query: 342 PDTKNKSTKKKLKKDDN-----------KPSEKALALEENVNRTEKKKKNRKNKEQSEFD 390
P K K ++ LK +P+E + N +T ++K +K KE E D
Sbjct: 296 PPPKTKPIEQTLKSGTKPSIAPARSGTKRPAENNDTKQSNKKKTTEEKNKKKEKEPEEDD 475
Query: 391 IIDDAEASFAEIFNIKDQENLNHG 414
++ +ASF +F D+ + G
Sbjct: 476 NSNNKKASFQRVFTEDDEVAILQG 547
>BG645847 similar to GP|11994124|d receptor protein kinase {Arabidopsis
thaliana}, partial (12%)
Length = 804
Score = 44.3 bits (103), Expect = 1e-04
Identities = 30/93 (32%), Positives = 47/93 (50%), Gaps = 2/93 (2%)
Frame = +2
Query: 143 KIGEALKKVKSITKLSLSHCQLEG-IDTSLKFCVELTELRLAHNDIKS-LPEELMHNSKL 200
K+G +L+K++ + LSLSH G I SL L +L L+HN LP ++ S +
Sbjct: 290 KLGRSLEKLQHLVTLSLSHNNFSGTISPSLTLSNTLQKLNLSHNSFSGPLPLSFVNMSSI 469
Query: 201 RNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGN 233
R +DL +N A ++ LR ++L N
Sbjct: 470 RFIDLSHNSFAGQMPDGFFENCFSLRRVSLSMN 568
>BF645340 weakly similar to GP|6016693|gb|A putative disease resistance
protein {Arabidopsis thaliana}, partial (7%)
Length = 655
Score = 43.9 bits (102), Expect = 1e-04
Identities = 56/217 (25%), Positives = 95/217 (42%), Gaps = 2/217 (0%)
Frame = +2
Query: 42 KLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMD 101
KLE LDL N L T + L++ +N S++ I ++ D
Sbjct: 8 KLESLDLSNNKLNGTVSNWLLETSRSLNLSQNLFTSIDQIS---------------RNSD 142
Query: 102 EIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSH 161
++G L L++ N VSICNL ++ LN + N I + L + S+ L L
Sbjct: 143 QLGDLDLSFNLLVG-NLSVSICNLSSLEFLN--LGHNNFTGNIPQCLANLPSLQILDLQM 313
Query: 162 CQLEG-IDTSLKFCVELTELRLAHNDIKS-LPEELMHNSKLRNLDLGNNVIAKWSDIKVL 219
G + + +L L L N ++ P+ L H L+ L+L NN + + L
Sbjct: 314 NNFYGTLPNNFSKSSKLITLNLNDNQLEGYFPKSLSHCENLQVLNLRNNKMEDKFPV-WL 490
Query: 220 KSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFN 256
++L L+ L L+ N + + + KI++ P L +F+
Sbjct: 491 QTLQYLKVLVLRDNKLHGHIANL-KIRHPFPSLVIFD 598
Score = 37.7 bits (86), Expect = 0.010
Identities = 37/129 (28%), Positives = 60/129 (45%), Gaps = 9/129 (6%)
Frame = +2
Query: 17 NPSSISSLHLTHKALSD--VSCLASFNKLEKLDLKFNNL--TSLEGLRACVTLKWLSVVE 72
N SS+ L+L H + CLA+ L+ LDL+ NN T L L++ +
Sbjct: 206 NLSSLEFLNLGHNNFTGNIPQCLANLPSLQILDLQMNNFYGTLPNNFSKSSKLITLNLND 385
Query: 73 NKLESL--EGIQGLTKLTVLNAGKNKLKSMDEI--GSLSTIRALILNDNEIVS-ICNLDQ 127
N+LE + + L VLN NK++ + +L ++ L+L DN++ I NL
Sbjct: 386 NQLEGYFPKSLSHCENLQVLNLRNNKMEDKFPVWLQTLQYLKVLVLRDNKLHGHIANLKI 565
Query: 128 MKELNTLVL 136
+LV+
Sbjct: 566 RHPFPSLVI 592
>TC87339 similar to PIR|D85041|D85041 hypothetical protein AT4g03260
[imported] - Arabidopsis thaliana, partial (40%)
Length = 1316
Score = 43.9 bits (102), Expect = 1e-04
Identities = 23/71 (32%), Positives = 41/71 (57%), Gaps = 1/71 (1%)
Frame = +2
Query: 38 ASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNK 96
+ +L LDL +N + + GL +C +LK L + NK+ +EG+ L KL++L+ NK
Sbjct: 347 SELTRLRVLDLSYNRILRIGHGLASCSSLKELYLAGNKIGEVEGLHRLLKLSILDIRFNK 526
Query: 97 LKSMDEIGSLS 107
+ + +G L+
Sbjct: 527 ISTAKCLGQLA 559
>TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F23H14.7
{Arabidopsis thaliana}, partial (43%)
Length = 1449
Score = 43.5 bits (101), Expect = 2e-04
Identities = 47/225 (20%), Positives = 91/225 (39%), Gaps = 3/225 (1%)
Frame = +3
Query: 243 RKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHDFSFDHVDQNEDDHLEAADKRKSNKK 302
+K+K A K + + D+D+ +E+ + +H H + DH + +K K
Sbjct: 798 KKLKTAKHKKRSGSDTDSDRDSDDERKRASKRSHRKHRKHSHIDSGDHEKRKEKSSKWKT 977
Query: 303 RKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKPSE 362
+K ++++S+ + D+ + ++ KKK + KK +D S
Sbjct: 978 KKRSSESSDFSS---DESESSSEEEKRRKKKQR--------------KKIRDQDSRSNSS 1106
Query: 363 KALALEENVNRTEKKKKNRKNKEQSEFDII-DDAEASFAEIFNIKDQENLNH--GGEMKL 419
+ + + VN+ +++ + R+ + S+ D D+ E S + + K + H E L
Sbjct: 1107 GSDSYADEVNKRKRRHRRRRPSKFSDSDFYSDEGECSDRKRGHGKHHKRHRHSESSESDL 1286
Query: 420 QDQVPKDLKLVSSIETLPVKHKSAKMHNVESLSSPGTEIGMGGPS 464
D+ S H+ + HN E SS G GG S
Sbjct: 1287 SSDESDDIHRKRSHRRHHKHHR--RSHNTEVRSSDSDYHGHGGRS 1415
>TC89762 weakly similar to GP|7268809|emb|CAB79014.1 leucine rich
repeat-like protein {Arabidopsis thaliana}, partial (6%)
Length = 1228
Score = 42.7 bits (99), Expect = 3e-04
Identities = 59/245 (24%), Positives = 103/245 (41%), Gaps = 40/245 (16%)
Frame = +3
Query: 29 KALSDVSCLASFNKLEKLDLKFNNLTSLEG------LRACVTLKWLSVVENKLE-SLEGI 81
K + D + + NK + + L F+ +SL G L+ L++ K + I
Sbjct: 192 KPICDFTGITCDNKGDIISLDFSGWSSLSGNFPSNICSYLPNLRVLNLGNTKFKFPTNSI 371
Query: 82 QGLTKLTVLNAGKNKLK-SMDEIGSLSTIRALILNDNEI-----VSICNLDQMKELNTLV 135
+ L +LN K L ++ + SL +R L L+ N +S+ NL ++ LN
Sbjct: 372 INCSHLELLNMNKMHLSGTLPDFSSLKYLRVLDLSYNSFTGDFPMSVFNLTNLEILNFNE 551
Query: 136 LSKNPIRKIGEALKKVKSITKLSLSHCQLEG-IDTSLKFCVELTELRLAHNDIK------ 188
SK + ++ ++ +++S+ + LS C L G I S+ L +L L+ N +
Sbjct: 552 NSKLNLWELPKSFVRLRSLKSMILSTCMLHGQIPPSISNITTLIDLELSGNFLTGQIPKE 731
Query: 189 --------------------SLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNL 228
S+PEEL + ++L +LD+ N + V K L KL+ L
Sbjct: 732 LGLLKNLQQLELYYNYFLVGSIPEELGNLTELVDLDMSVNKLTGTIPSSVCK-LPKLQVL 908
Query: 229 NLQGN 233
N
Sbjct: 909 QFYNN 923
>AW685527 similar to GP|1946300|emb hypothetical protein {Silene latifolia},
partial (19%)
Length = 403
Score = 42.4 bits (98), Expect = 4e-04
Identities = 27/97 (27%), Positives = 50/97 (50%), Gaps = 1/97 (1%)
Frame = +3
Query: 134 LVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEE 193
L+L+ N I + E ++ + + L+LSH L + ++ EL L ++HN I +PEE
Sbjct: 96 LILAHNSIELLKEDIRNLSCLVVLNLSHNSLSQLPAAIGELPELKMLDVSHNLIVRIPEE 275
Query: 194 LMHNSKLRNLDLGNNVIAKW-SDIKVLKSLTKLRNLN 229
+ + L D NN + + S++ +L+ L+ N
Sbjct: 276 IGSAASLVKFDCSNNQLTELPSELGRCLALSDLKGSN 386
Score = 33.9 bits (76), Expect = 0.14
Identities = 17/58 (29%), Positives = 32/58 (54%)
Frame = +3
Query: 155 TKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAK 212
+KL L+H +E + ++ L L L+HN + LP + +L+ LD+ +N+I +
Sbjct: 90 SKLILAHNSIELLKEDIRNLSCLVVLNLSHNSLSQLPAAIGELPELKMLDVSHNLIVR 263
>TC81060 similar to GP|22136192|gb|AAM91174.1 unknown protein {Arabidopsis
thaliana}, partial (95%)
Length = 1114
Score = 41.2 bits (95), Expect = 9e-04
Identities = 31/82 (37%), Positives = 44/82 (52%), Gaps = 6/82 (7%)
Frame = +1
Query: 84 LTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEI----VSICNLDQMKELNTLVLSK 138
L L +LN NKL S+ E +GS ++ L NDN I S+CNL +K +L L
Sbjct: 451 LRNLLILNISNNKLHSLPESVGSCFSLEELQANDNLIEDLPSSVCNLSHLK---SLCLDT 621
Query: 139 NPIRKIG-EALKKVKSITKLSL 159
N +++I LK K++ LSL
Sbjct: 622 NNVKQIPMNLLKDCKALQNLSL 687
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,163,333
Number of Sequences: 36976
Number of extensions: 149908
Number of successful extensions: 1075
Number of sequences better than 10.0: 163
Number of HSP's better than 10.0 without gapping: 1012
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1048
length of query: 468
length of database: 9,014,727
effective HSP length: 100
effective length of query: 368
effective length of database: 5,317,127
effective search space: 1956702736
effective search space used: 1956702736
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146332.10


