

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146331.8 + phase: 2 /pseudo
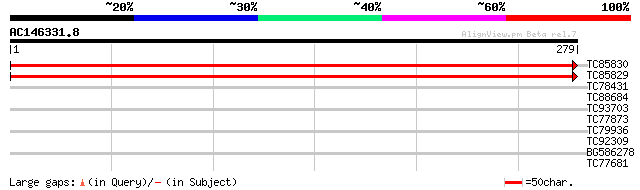
(279 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85830 weakly similar to GP|16323424|gb|AAL15206.1 unknown prot... 545 e-156
TC85829 weakly similar to GP|16323424|gb|AAL15206.1 unknown prot... 525 e-149
TC78431 similar to GP|16797791|gb|AAL27150.1 bZIP transcription ... 31 0.60
TC88684 similar to GP|7576200|emb|CAB87861.1 protein synthesis i... 30 1.3
TC93703 similar to GP|9294064|dbj|BAB02021.1 DNA-dependent RNA p... 28 3.0
TC77873 similar to GP|14517486|gb|AAK62633.1 AT5g20650/T1M15_50 ... 28 3.0
TC79936 similar to PIR|T51453|T51453 serine/threonine specific p... 28 3.9
TC92309 similar to PIR|T04107|T04107 calmodulin-binding heat-sho... 27 6.6
BG586278 homologue to PIR|E86217|E86 protein T27G7.10 [imported]... 27 8.7
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 27 8.7
>TC85830 weakly similar to GP|16323424|gb|AAL15206.1 unknown protein
{Arabidopsis thaliana}, partial (43%)
Length = 1344
Score = 545 bits (1403), Expect = e-156
Identities = 279/279 (100%), Positives = 279/279 (100%)
Frame = +2
Query: 1 VSENGLKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL 60
VSENGLKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL
Sbjct: 233 VSENGLKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL 412
Query: 61 SLNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA 120
SLNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA
Sbjct: 413 SLNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA 592
Query: 121 FLGSCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGE 180
FLGSCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGE
Sbjct: 593 FLGSCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGE 772
Query: 181 VLSELISSVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQ 240
VLSELISSVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQ
Sbjct: 773 VLSELISSVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQ 952
Query: 241 LKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ 279
LKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ
Sbjct: 953 LKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ 1069
>TC85829 weakly similar to GP|16323424|gb|AAL15206.1 unknown protein
{Arabidopsis thaliana}, partial (42%)
Length = 1353
Score = 525 bits (1351), Expect = e-149
Identities = 268/279 (96%), Positives = 272/279 (97%)
Frame = +2
Query: 1 VSENGLKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL 60
+S+NGLKESVNYQVIS KDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL
Sbjct: 257 ISDNGLKESVNYQVISPKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL 436
Query: 61 SLNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA 120
SLNKHADSALS LKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA
Sbjct: 437 SLNKHADSALSDFLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA 616
Query: 121 FLGSCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGE 180
FLGSCSMG GNREET AL SVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVD T+SEGE
Sbjct: 617 FLGSCSMGTGNREETKALHSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDRTQSEGE 796
Query: 181 VLSELISSVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQ 240
VLSELISSVEESGAKYAVLYVSD+SRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQ
Sbjct: 797 VLSELISSVEESGAKYAVLYVSDLSRSIQYPSYRDLQRFLAESTTGNGSTNSTACDGVCQ 976
Query: 241 LKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ 279
LKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ
Sbjct: 977 LKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ 1093
>TC78431 similar to GP|16797791|gb|AAL27150.1 bZIP transcription factor
{Nicotiana tabacum}, partial (61%)
Length = 1546
Score = 30.8 bits (68), Expect = 0.60
Identities = 25/79 (31%), Positives = 36/79 (44%), Gaps = 10/79 (12%)
Frame = -3
Query: 60 LSLNKHADSALSYLLKDSFVRSNTSMAFPYV----SASEDVNLEDSLVSG------FAEA 109
L L + ADS L +L F + ++ YV S+S D +L+D LV G F
Sbjct: 806 LLLERLADSLLESILLTRFTSVGSVLSIIYVSPSASSSSDCSLDDPLVDGLIATMVFFCT 627
Query: 110 CGDDLGIGNVAFLGSCSMG 128
CG + G SC++G
Sbjct: 626 CGMEGTAGGTLASLSCNIG 570
>TC88684 similar to GP|7576200|emb|CAB87861.1 protein synthesis initiation
factor-like {Arabidopsis thaliana}, partial (3%)
Length = 1322
Score = 29.6 bits (65), Expect = 1.3
Identities = 16/40 (40%), Positives = 25/40 (62%), Gaps = 3/40 (7%)
Frame = -3
Query: 68 SALSYLLKDSFVRSNTSMAFPYVSASED---VNLEDSLVS 104
+ LS+LLK++F+ SNT + P +SA + L DS+ S
Sbjct: 813 TTLSFLLKETFLESNTEVDMPKLSAIRSQSVLQLADSISS 694
>TC93703 similar to GP|9294064|dbj|BAB02021.1 DNA-dependent RNA polymerase
II {Arabidopsis thaliana}, partial (17%)
Length = 598
Score = 28.5 bits (62), Expect = 3.0
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +1
Query: 50 FVGGELQSSDLSLNKHADSALSYLLK 75
FVGG LQS D+ + + ADS + +K
Sbjct: 55 FVGGNLQSGDIIIGRCADSGTDHSIK 132
>TC77873 similar to GP|14517486|gb|AAK62633.1 AT5g20650/T1M15_50
{Arabidopsis thaliana}, partial (66%)
Length = 732
Score = 28.5 bits (62), Expect = 3.0
Identities = 11/22 (50%), Positives = 18/22 (81%)
Frame = +1
Query: 245 LLEGLLVGIVLLIILISGLCCM 266
LL GLL+ I+++II+I +CC+
Sbjct: 574 LLMGLLICIIIIIIIIPKVCCI 639
>TC79936 similar to PIR|T51453|T51453 serine/threonine specific protein
kinase-like - Arabidopsis thaliana, partial (22%)
Length = 869
Score = 28.1 bits (61), Expect = 3.9
Identities = 19/69 (27%), Positives = 32/69 (45%)
Frame = +1
Query: 154 KGKTDLVVFCNGPQASKNVDSTKSEGEVLSELISSVEESGAKYAVLYVSDISRSIQYPSY 213
+ K D + CNG +++N D+TKS + SE S E+ K ++ V + + S
Sbjct: 397 RSKLDRSICCNGNTSTQNGDNTKSV-IIQSEENKSTSETNTKESIGLVDSSTTTSNGESI 573
Query: 214 RDLQRFLAE 222
+F E
Sbjct: 574 SSTSKFSEE 600
>TC92309 similar to PIR|T04107|T04107 calmodulin-binding heat-shock protein
- common tobacco, partial (36%)
Length = 763
Score = 27.3 bits (59), Expect = 6.6
Identities = 17/62 (27%), Positives = 26/62 (41%), Gaps = 11/62 (17%)
Frame = -2
Query: 216 LQRFLAESTTGNGSTNSTACDGVCQLK-----------SSLLEGLLVGIVLLIILISGLC 264
L+ FL T C G C LK SL+EGL + + +++ G+C
Sbjct: 228 LESFLRSDTEFIKKKKVDVCWGSC*LKVDLV*EWFS*RESLMEGLSIC*IAKVVMSCGMC 49
Query: 265 CM 266
C+
Sbjct: 48 CL 43
>BG586278 homologue to PIR|E86217|E86 protein T27G7.10 [imported] -
Arabidopsis thaliana, partial (8%)
Length = 755
Score = 26.9 bits (58), Expect = 8.7
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = +1
Query: 193 GAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNG 228
G + YV+D+S SIQ +++ R L S +G G
Sbjct: 40 GKTMLLYYVNDVSLSIQNDNFQSALRVLMNSLSGAG 147
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 26.9 bits (58), Expect = 8.7
Identities = 16/45 (35%), Positives = 21/45 (46%)
Frame = +2
Query: 140 SVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSE 184
S+ L K E SH K V FC ++ D ++ EGE SE
Sbjct: 275 SLDIVLEKEFELSHNSKAASVHFCGYKAYYEDNDDSEEEGETDSE 409
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,377,919
Number of Sequences: 36976
Number of extensions: 88724
Number of successful extensions: 362
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 362
length of query: 279
length of database: 9,014,727
effective HSP length: 95
effective length of query: 184
effective length of database: 5,502,007
effective search space: 1012369288
effective search space used: 1012369288
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146331.8


