

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145513.6 + phase: 0
(882 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
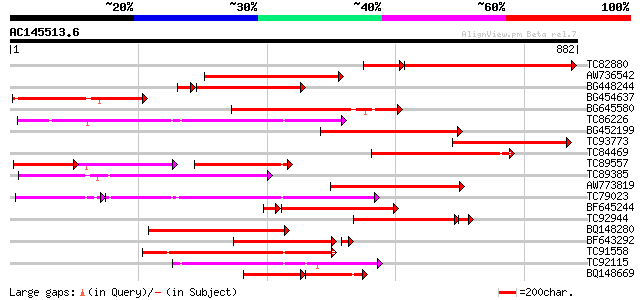
Score E
Sequences producing significant alignments: (bits) Value
TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein ... 456 e-159
AW736542 similar to PIR|T07656|T076 probable resistance protein ... 350 1e-96
BG448244 weakly similar to PIR|T08833|T088 disease resistance pr... 295 4e-89
BG454637 weakly similar to GP|23477203|emb TIR-NBS disease resis... 315 6e-86
BG645580 similar to PIR|T07656|T076 probable resistance protein ... 289 3e-78
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 286 2e-77
BG452199 weakly similar to GP|10178211|db TMV resistance protein... 279 4e-75
TC93773 weakly similar to GP|9858483|gb|AAG01054.1| resistance p... 278 7e-75
TC84469 similar to GP|9858483|gb|AAG01054.1| resistance protein ... 263 2e-70
TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 111 1e-66
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 248 7e-66
AW773819 similar to GP|9858483|gb|A resistance protein MG55 {Gly... 246 3e-65
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 236 2e-62
BF645244 weakly similar to PIR|T07656|T076 probable resistance p... 219 5e-62
TC92944 similar to GP|9858483|gb|AAG01054.1| resistance protein ... 209 1e-58
BQ148280 similar to GP|7107246|gb| unknown {Cicer arietinum}, pa... 218 6e-57
BF643292 weakly similar to GP|22037340|gb| disease resistance-li... 205 7e-55
TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll... 211 1e-54
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 209 4e-54
BQ148669 similar to PIR|T07656|T076 probable resistance protein ... 119 1e-50
>TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (46%)
Length = 1238
Score = 456 bits (1174), Expect(2) = e-159
Identities = 234/268 (87%), Positives = 248/268 (92%)
Frame = +2
Query: 614 LIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLKNLLMLNLKDC 673
LIEGLKILNLSHSK LKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDL NLL++NLKDC
Sbjct: 296 LIEGLKILNLSHSKYLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLNNLLLINLKDC 475
Query: 674 TSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRS 733
TSL NLPREIYQLR V+TLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRS
Sbjct: 476 TSLSNLPREIYQLRTVKTLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRS 655
Query: 734 KSIGYISLCGYEGLSHHVFPSLIRSWISPTMNSLPRIPPFGGMSKSLFSLDIDSNNLALV 793
KSIGYISLCGYEGLSHHVFPSLIRSW+SPTMNS+ I PFGGMSKSL SLDI+SNNLALV
Sbjct: 656 KSIGYISLCGYEGLSHHVFPSLIRSWMSPTMNSVAHISPFGGMSKSLASLDIESNNLALV 835
Query: 794 SQSQILNSCSRLRSVSVQCDSEIQLKQEFGRFLDDLYDAGLTEMRTSHALQISNLTMRSL 853
QSQIL+SCS+LRSVSVQCDSEIQLKQEF RFLDDLYDAGLTE+ SHA IS+ ++RSL
Sbjct: 836 YQSQILSSCSKLRSVSVQCDSEIQLKQEFRRFLDDLYDAGLTELGISHASHISDHSLRSL 1015
Query: 854 LFGIGSCHIVINTLRKSLSQVSSLSSHL 881
L G+G+CHIVIN L KSLSQV S SSHL
Sbjct: 1016LIGMGNCHIVINILGKSLSQVPSQSSHL 1099
Score = 124 bits (311), Expect(2) = e-159
Identities = 55/64 (85%), Positives = 60/64 (92%)
Frame = +3
Query: 551 RLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKHSNLAQVWK 610
+LRLLQLDNVQVIGDY+CF K LRWLSWQGFPLKY PENFYQKN+VAM+LKHSNL QVWK
Sbjct: 3 KLRLLQLDNVQVIGDYECFSKQLRWLSWQGFPLKYMPENFYQKNVVAMDLKHSNLTQVWK 182
Query: 611 KPQL 614
KPQ+
Sbjct: 183 KPQV 194
>AW736542 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (83%)
Length = 656
Score = 350 bits (898), Expect = 1e-96
Identities = 175/218 (80%), Positives = 196/218 (89%), Gaps = 1/218 (0%)
Frame = +2
Query: 303 DALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPT 361
+ALC RN +GPGSIIIITTRD R+L+IL VDFIYEAE LN ES LF HAF++A P+
Sbjct: 2 NALCGNRNGIGPGSIIIITTRDARLLDILGVDFIYEAEGLNVHESRRLFNWHAFKEANPS 181
Query: 362 QDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFD 421
+ FLILS DVV+YCGG+PLALEVLGSYLF R+K+EW+SV+SKL+KIPNDQIHE LKISFD
Sbjct: 182 EAFLILSGDVVSYCGGLPLALEVLGSYLFNRRKREWQSVISKLQKIPNDQIHEKLKISFD 361
Query: 422 GLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGM 481
GL+D MEKNIFLDVCCFFIGKDRAYVT+ILNGCGL+ADIGI VLIERSL+KVEKN KLGM
Sbjct: 362 GLEDHMEKNIFLDVCCFFIGKDRAYVTEILNGCGLHADIGIEVLIERSLLKVEKNNKLGM 541
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLAD 519
HALLRDMGREIVRESSPEEPEK TRLWC EDVV+VLA+
Sbjct: 542 HALLRDMGREIVRESSPEEPEKRTRLWCFEDVVDVLAE 655
>BG448244 weakly similar to PIR|T08833|T088 disease resistance protein
homolog RLG6 - soybean (fragment), partial (34%)
Length = 599
Score = 295 bits (756), Expect(2) = 4e-89
Identities = 148/169 (87%), Positives = 157/169 (92%)
Frame = +2
Query: 291 AVLDDVSELEQFDALCQRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELF 350
AVLDDVSELEQF+ALC+ NSVGPGS+IIITTRDLRVLNILEVDFIYEAE LNASESLELF
Sbjct: 92 AVLDDVSELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELF 271
Query: 351 CKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPND 410
C HAFRK IPT+DFLILSR VVAYCGGIPLALEVLGSYL KR+KQEW+SVLSKLEKIPND
Sbjct: 272 CGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPND 451
Query: 411 QIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNAD 459
QIHE LKI F+GL DRME +IFLDVCCFFIGKDR YVTKILNG GL+AD
Sbjct: 452 QIHEKLKIXFNGLSDRMEXDIFLDVCCFFIGKDRXYVTKILNGXGLHAD 598
Score = 52.4 bits (124), Expect(2) = 4e-89
Identities = 26/29 (89%), Positives = 28/29 (95%)
Frame = +1
Query: 261 DILKTRKIKVLSVEQGKTMIKQQLRAKRI 289
DILKTRKIKVLSVEQGK MIKQ+LR+KRI
Sbjct: 1 DILKTRKIKVLSVEQGKAMIKQRLRSKRI 87
>BG454637 weakly similar to GP|23477203|emb TIR-NBS disease resistance
protein {Populus balsamifera subsp. trichocarpa},
partial (16%)
Length = 659
Score = 315 bits (806), Expect = 6e-86
Identities = 159/214 (74%), Positives = 183/214 (85%), Gaps = 4/214 (1%)
Frame = +3
Query: 5 RSKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRA 64
RS+PQ W +DVFINFRG DTRKTF+SHLY ALT+AGINTFLD+ENL+KG+ELGPEL+RA
Sbjct: 27 RSRPQ--WIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPELVRA 200
Query: 65 IQGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTRFS 124
IQGSQIAIVVFSKNYV+S WCL+EL+QIM+CKA++GQVVMPVF ITPSNIRQ++
Sbjct: 201 IQGSQIAIVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHS----- 365
Query: 125 ETTLFFDELVPFM----NTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLPLP 180
+ DEL + L+D SYL+GWD+SNYSN+SKVVKEIVSQVLKNLD KYLPLP
Sbjct: 366 -PVILVDELDQIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLP 542
Query: 181 DFQVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGG 214
+FQVGL+PRAEK IRFLRQNTR VCLVGIWGMGG
Sbjct: 543 NFQVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGG 644
>BG645580 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (87%)
Length = 814
Score = 289 bits (740), Expect = 3e-78
Identities = 155/280 (55%), Positives = 194/280 (68%), Gaps = 13/280 (4%)
Frame = +1
Query: 345 ESLELFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKL 404
+ LELF HAFR+ P ++F LSR VVAYCGG+PLALEV+GSYL+ R KQEW SVL KL
Sbjct: 1 DPLELFSWHAFRQPSPIKNFSELSRTVVAYCGGLPLALEVIGSYLYGRTKQEWESVLLKL 180
Query: 405 EKIPNDQIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITV 464
E+IPNDQ+ E L+IS+DGLKD M K+IFLD+CCFFIGKDRAYVT+ILNGCGL ADIGITV
Sbjct: 181 ERIPNDQVQEKLRISYDGLKDDMAKDIFLDICCFFIGKDRAYVTEILNGCGLYADIGITV 360
Query: 465 LIERSLIKVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTK 524
L+ERSL+K+EKN KLGMH LLRDMGREIVR+SS + P K +RLW HEDV +VL TGT
Sbjct: 361 LVERSLVKIEKNNKLGMHDLLRDMGREIVRQSSAKNPGKRSRLWFHEDVHDVLTKNTGTI 540
Query: 525 AIEGLVMKLPKTNRVCFDTIAFEKMIRL-------------RLLQLDNVQVIGDYKCFPK 571
+EG+V VC + F ++ L L+ L +++++G FP
Sbjct: 541 NVEGVVF------*VCQEPAEFASVLILSWK*NN*NN*NFYNLIVLTSLEIMG---VFPN 693
Query: 572 HLRWLSWQGFPLKYTPENFYQKNLVAMELKHSNLAQVWKK 611
+ + P F +KNLVA++LKHS + QVW +
Sbjct: 694 N*DGSVCKDLP*TVYQMTFIRKNLVALDLKHSKIKQVWNE 813
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 286 bits (733), Expect = 2e-77
Identities = 189/530 (35%), Positives = 303/530 (56%), Gaps = 17/530 (3%)
Frame = +3
Query: 12 WTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNEN--LQKGKELGPELIRAIQGSQ 69
++YDVF+ +G DTR F +L AL + GI TF D+++ LQ+ ++ P +I + S+
Sbjct: 84 FSYDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDKVTPIII---EESR 254
Query: 70 IAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYA---------- 119
I I +FS NY S CL L I+ C G +V+PVF+ + P+++R +
Sbjct: 255 ILIPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVRHHTGRYGKALAEH 434
Query: 120 VTRFSETTLFFDELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLPL 179
RF T + L + L A+ L + ++ E +++ +IV + + + L +
Sbjct: 435 ENRFQNDTKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGKIVKYISNKISRQSLHV 614
Query: 180 PDFQVGLEPRAEKSIRFLRQNTR-GVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFL 238
+ VGL+ R ++ L + GV +VGI+G+GG GKST+A+ IYN + +FE FL
Sbjct: 615 ATYPVGLQSRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLARAIYNFVADQFEGLCFL 794
Query: 239 ANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSE 298
+RE + + QE LS L+ KIK+ V +G ++IK++L K+IL +LDDV
Sbjct: 795 EQVRENSASNSLK-RFQEMLLSKTLQL-KIKLADVSEGISIIKERLCRKKILLILDDVDN 968
Query: 299 LEQFDALCQR-NSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRK 357
++Q +AL + GPGS +IITTRD +L E++ Y + LN +E+LEL AF+
Sbjct: 969 MKQLNALAGGVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNVTEALELLRWMAFKN 1148
Query: 358 -AIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEIL 416
+P+ IL+R VVAY G+P+ +E++GS LF + +E ++ L EKIPN +I IL
Sbjct: 1149DKVPSSYEKILNR-VVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNKEIQRIL 1325
Query: 417 KISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNG-CGLNADIGITVLIERSLI-KVE 474
K+S+D L++ E+++FLD+ C F G V +IL+ G + + VL+E+ LI E
Sbjct: 1326KVSYDSLEEE-EQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHVEVLVEKCLIDHFE 1502
Query: 475 KNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTK 524
+ + +H L+ +MG+E+VR SP EP K +RLW +D+ VL + T +K
Sbjct: 1503YDSHVSLHNLIENMGKELVRLESPFEPGKRSRLWFEKDIFEVLEENTVSK 1652
>BG452199 weakly similar to GP|10178211|db TMV resistance protein N
{Arabidopsis thaliana}, partial (4%)
Length = 666
Score = 279 bits (713), Expect = 4e-75
Identities = 135/221 (61%), Positives = 174/221 (78%)
Frame = +3
Query: 484 LLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCFDT 543
LLRDMGREIVRE SP+EPE+ +RLW ++DV++VL++ TGTKA+EGL +KLP N CF T
Sbjct: 3 LLRDMGREIVREKSPKEPEERSRLWFNKDVIDVLSEKTGTKAVEGLALKLPTANGKCFST 182
Query: 544 IAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKHS 603
AF++M +LRLLQL +VQ+ GD++ ++L WLSW GFPLK P +FYQ+NLV++EL +S
Sbjct: 183 KAFKEMKKLRLLQLASVQLEGDFEYLSRNLIWLSWNGFPLKCIPSSFYQENLVSIELVNS 362
Query: 604 NLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLK 663
N+ VWK+PQ +E LKILNLSHS L +TPDFS LPNLE+L++ DC L EV SIGDL
Sbjct: 363 NVKVVWKEPQRLEKLKILNLSHSHFLSKTPDFSNLPNLEQLVLTDCPKLSEVSQSIGDLN 542
Query: 664 NLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLE 704
+L++NL+DC SL +LPR IY L+ + TLILSGC IDKLE
Sbjct: 543 KILLINLEDCISLQSLPRSIYNLKSLNTLILSGCLMIDKLE 665
>TC93773 weakly similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (31%)
Length = 856
Score = 278 bits (711), Expect = 7e-75
Identities = 143/184 (77%), Positives = 158/184 (85%)
Frame = +2
Query: 690 ETLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEGLSH 749
E ILSGCSKI+KLEEDIVQM+SLTTL+AA TGVKQ PFSIV+SK+IGYISLC YEGLS
Sbjct: 17 ENFILSGCSKIEKLEEDIVQMKSLTTLIAAKTGVKQVPFSIVKSKNIGYISLCEYEGLSR 196
Query: 750 HVFPSLIRSWISPTMNSLPRIPPFGGMSKSLFSLDIDSNNLALVSQSQILNSCSRLRSVS 809
VFPS+I SW+SP MNSL IPP GGMS SL LD+DS NL LV QS IL+S S+LRSVS
Sbjct: 197 DVFPSIIWSWMSPNMNSLAHIPPVGGMSMSLVCLDVDSRNLGLVHQSPILSSYSKLRSVS 376
Query: 810 VQCDSEIQLKQEFGRFLDDLYDAGLTEMRTSHALQISNLTMRSLLFGIGSCHIVINTLRK 869
VQCDSEIQLKQEF RFLDD+YDAGLTE TSH QI +L++RSLLFGIGSCHIVINTLRK
Sbjct: 377 VQCDSEIQLKQEFRRFLDDIYDAGLTEFGTSHGSQILDLSLRSLLFGIGSCHIVINTLRK 556
Query: 870 SLSQ 873
SLS+
Sbjct: 557 SLSE 568
>TC84469 similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (39%)
Length = 735
Score = 263 bits (672), Expect = 2e-70
Identities = 133/221 (60%), Positives = 168/221 (75%)
Frame = +3
Query: 564 GDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKHSNLAQVWKKPQLIEGLKILNL 623
GD+K ++L+WL W GFPL+ P NFYQ+N+V++EL++SN VWK+ Q +E LKILNL
Sbjct: 27 GDFKYISRNLKWLHWNGFPLRCIPSNFYQRNIVSIELENSNAKLVWKEIQRMEQLKILNL 206
Query: 624 SHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLKNLLMLNLKDCTSLGNLPREI 683
SHS +L +TPDFS LPNLEKL+++DC L +V SIG LK ++++NLKDC SL +LPR I
Sbjct: 207 SHSHHLTQTPDFSYLPNLEKLVLEDCPRLSQVSHSIGHLKKVVLINLKDCISLCSLPRNI 386
Query: 684 YQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCG 743
Y L+ + TLILSGC IDKLEED+ QMESLTTL+A NTG+ + PFS+VRSKSIG+ISLCG
Sbjct: 387 YTLKTLNTLILSGCLMIDKLEEDLEQMESLTTLIANNTGITKVPFSLVRSKSIGFISLCG 566
Query: 744 YEGLSHHVFPSLIRSWISPTMNSLPRIPPFGGMSKSLFSLD 784
YEG S VFPS+I SW+SP N P MS SL SL+
Sbjct: 567 YEGFSRDVFPSIIWSWMSPN-NLSPAFQTASHMS-SLVSLE 683
>TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein N
{Arabidopsis thaliana}, partial (8%)
Length = 1567
Score = 111 bits (277), Expect(3) = 1e-66
Identities = 61/153 (39%), Positives = 94/153 (60%), Gaps = 1/153 (0%)
Frame = +3
Query: 288 RILAVLDDVSELEQFDAL-CQRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASES 346
++L VLD+V +LEQ + L + + P S III TRD +L D ++EA+ +N ++
Sbjct: 1110 KLLIVLDNVEQLEQLEKLDIEPKFLHPRSKIIIITRDKHILQAYGADEVFEAKLMNDEDA 1289
Query: 347 LELFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEK 406
+L C+ AF+ P+ F L V+ Y +PLA++VLGS+LF R EW S L K EK
Sbjct: 1290 HKLLCRKAFKSDYPSSGFAELIPKVLVYAQRLPLAVKVLGSFLFSRNANEWSSTLDKFEK 1469
Query: 407 IPNDQIHEILKISFDGLKDRMEKNIFLDVCCFF 439
P ++I + L++S++GL ++ EK +FL V FF
Sbjct: 1470 NPPNKIMKALQVSYEGL-EKDEKEVFLHVAXFF 1565
Score = 104 bits (260), Expect(3) = 1e-66
Identities = 50/102 (49%), Positives = 67/102 (65%)
Frame = +3
Query: 6 SKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAI 65
S + YDVFI+FRG+DTR +F+ HLY L GI TF D++ LQKG+ + P+L++AI
Sbjct: 240 SNSNQSYRYDVFISFRGSDTRNSFVDHLYAHLNRKGIFTFKDDKQLQKGEAISPQLLQAI 419
Query: 66 QGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVF 107
Q S+I+IVVFSK+Y S WCL E+ I E + N P F
Sbjct: 420 QQSRISIVVFSKDYASSTWCLDEMAAINESRINLKTSCFPCF 545
Score = 78.2 bits (191), Expect(3) = 1e-66
Identities = 47/170 (27%), Positives = 90/170 (52%), Gaps = 9/170 (5%)
Frame = +1
Query: 101 QVVMPVFYCITPSNIRQ----YAVTRFSETTLF---FDELVPFMNTLQDASYLSGWDLSN 153
QVV PVFY + PS++++ Y T F ++ + T+ +GWD+
Sbjct: 526 QVVFPVFYDVDPSHVKKQNGVYENAFVLHTEAFKHNSGKVARWRTTMTYLGGTAGWDV-R 702
Query: 154 YSNESKVVKEIVSQVLKNLDNKYLPLPDFQVGLEPRAEKSIRFLRQNTR--GVCLVGIWG 211
E +++++IV V+K L + D +G++P E L+ +++ G ++GIWG
Sbjct: 703 VEPEFEMIEKIVEAVIKKLGRTFSGSTDDLIGIQPHIEALENLLKLSSKDDGCRVLGIWG 882
Query: 212 MGGIGKSTIAKVIYNDLCYEFENQSFLANIREVWEKDRGRIDLQEQFLSD 261
M GIGK+T+A V+Y+ + ++F+ F+ N+ +++E + +LS+
Sbjct: 883 MDGIGKTTLATVLYDKISFQFDACCFIENVSKIYEDGGACCCAETNYLSN 1032
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 248 bits (633), Expect = 7e-66
Identities = 139/413 (33%), Positives = 232/413 (55%), Gaps = 17/413 (4%)
Frame = +3
Query: 14 YDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQIAIV 73
YDVF+ FRG DTR F + L+ AL GI F D+ NL KG+ +GPEL+R I+GSQ+ +
Sbjct: 69 YDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRTIEGSQVFVA 248
Query: 74 VFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTRFSETTLFFDEL 133
V S+NY S WCL EL++I EC G+ V+P+FY + PS +++ ++ +++D+
Sbjct: 249 VLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKK-------QSGIYWDDF 407
Query: 134 VP--------------FMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLPL 179
+ L ++GWDL + +S V++IV +L L K +
Sbjct: 408 AKHEQRFKQDPHKVSRWREALNQVGSIAGWDLRD-KQQSVEVEKIVQTILNILKCKSSFV 584
Query: 180 PDFQVGLEPRAEK-SIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFL 238
VG+ R E + L + GV ++GIWGMGG GK+T+A +Y +C+ F+ F+
Sbjct: 585 SKDLVGINSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFI 764
Query: 239 ANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSE 298
++ +++ G ID Q+Q L L ++ + +I+ +L ++ L +LD+V +
Sbjct: 765 DDVSKIFRLHDGPIDAQKQILHQTLGIEHHQICNHYSATDLIRHRLSREKTLLILDNVDQ 944
Query: 299 LEQFDAL-CQRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFR- 356
+EQ + + R +G GS I+I +RD +L +VD Y+ L+ +ES +LF + AF+
Sbjct: 945 VEQLERIGVHREWLGAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAFKL 1124
Query: 357 KAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPN 409
+ I +++ L+ +++ Y G+PL + VLGS+L R EW+S L++L + PN
Sbjct: 1125EKIIMKNYQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRPN 1283
>AW773819 similar to GP|9858483|gb|A resistance protein MG55 {Glycine max},
partial (20%)
Length = 633
Score = 246 bits (628), Expect = 3e-65
Identities = 120/209 (57%), Positives = 156/209 (74%)
Frame = +1
Query: 499 EEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCFDTIAFEKMIRLRLLQLD 558
EEPE +RLW +DV +VL++ GTK +EGL +KLP+ N CF T AF+KM +LRLLQL
Sbjct: 4 EEPEGRSRLWFDKDVFDVLSEQNGTKVVEGLALKLPRENAKCFSTKAFKKMEKLRLLQLA 183
Query: 559 NVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKHSNLAQVWKKPQLIEGL 618
VQ+ GD++ ++LRWLSW GFPL P +FYQ NLV++EL +SN+ VWKK Q +E L
Sbjct: 184 GVQLDGDFEHLSRNLRWLSWNGFPLTCIPSSFYQGNLVSIELVNSNIKLVWKKTQRLEKL 363
Query: 619 KILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLKNLLMLNLKDCTSLGN 678
KILNLSHS L +TPDFS LPNLE+L++ DC L EV SI L +L++NL+DC L +
Sbjct: 364 KILNLSHSHYLTQTPDFSNLPNLEQLVLTDCPRLSEVSHSIEHLNKILLINLEDCIGLQS 543
Query: 679 LPREIYQLRRVETLILSGCSKIDKLEEDI 707
LPR IY+L+ ++TLILSGCS IDK+EED+
Sbjct: 544 LPRSIYKLKSLKTLILSGCSMIDKVEEDL 630
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 236 bits (603), Expect = 2e-62
Identities = 160/432 (37%), Positives = 253/432 (58%), Gaps = 7/432 (1%)
Frame = +3
Query: 150 DLSNYSNESKVVKEIVSQVLKNLDNKYLPLPDFQVGLEPRAEKSIRFLRQNTRG-VCLVG 208
D++ Y E + + +IV + + + L + + VGL+ R + L + + V +VG
Sbjct: 678 DMNRY--EYEFIGKIVEDISNRISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVG 851
Query: 209 IWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIREVWEKDRGRIDLQEQFLSDILKTRK- 267
++G GGIGKST+AK IYN + +FE FL N+R D + LQE+ L LKT +
Sbjct: 852 LYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLK-HLQEKLL---LKTVRL 1019
Query: 268 -IKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFDALCQR-NSVGPGSIIIITTRDLR 325
IK+ V QG +IKQ+L K+IL +LDDV +L+Q +AL + GPGS +IITTR+
Sbjct: 1020 DIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKH 1199
Query: 326 VLNILEVDFIYEAEELNASESLELFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVL 385
+L I ++ + E LNA+E+LEL AF++ +P+ IL+R + Y G+PLA+ ++
Sbjct: 1200 LLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNR-ALTYASGLPLAIVII 1376
Query: 386 GSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRA 445
GS L R Q+ S L E+IPN +I ILK+S+D L ++ E+++FLD+ C F G
Sbjct: 1377 GSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDSL-EKEEQSVFLDIACCFKGCKWP 1553
Query: 446 YVTKILNGCGLNADIG-ITVLIERSLIK-VEKNKKLGMHALLRDMGREIVRESSPEEPEK 503
V +IL+ + + + VL E+SL+ ++ + + +H L+ DMG+E+VR+ SP+EP +
Sbjct: 1554 EVKEILHAHYGHCIVHHVAVLAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEPGE 1733
Query: 504 HTRLWCHEDVVNVLADYTGTKAIEGLVMKLPK-TNRVCFDTIAFEKMIRLRLLQLDNVQV 562
+RLW D+V+VL TGT+ I+ + MK P + + ++ AFEKM L+ +N
Sbjct: 1734 RSRLWFERDIVHVLKKNTGTRKIKMINMKFPSMESDIDWNGNAFEKMTNLKTFITENGHH 1913
Query: 563 IGDYKCFPKHLR 574
+ P LR
Sbjct: 1914 SKSLEYLPSSLR 1949
Score = 109 bits (272), Expect = 5e-24
Identities = 59/140 (42%), Positives = 83/140 (59%)
Frame = +1
Query: 10 SKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQ 69
S +TY VF++FRGADTR F +LY ALT+ GI TF+D+ +LQ+G E+ P L AI+ S+
Sbjct: 82 SSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSR 261
Query: 70 IAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTRFSETTLF 129
I I VFS+NY S +CL EL I C G +V+PVF + P+++R + R+ E
Sbjct: 262 IFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHH-TGRYGEALAV 438
Query: 130 FDELVPFMNTLQDASYLSGW 149
+ F N + L W
Sbjct: 439 HKK--KFQNDKDNTERLQQW 492
>BF645244 weakly similar to PIR|T07656|T076 probable resistance protein -
soybean (fragment), partial (81%)
Length = 678
Score = 219 bits (558), Expect(2) = 5e-62
Identities = 107/183 (58%), Positives = 135/183 (73%)
Frame = +3
Query: 423 LKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGMH 482
L D EK IFLDV CFFIG DR VT ILNGC L +IGI++L+ERSL+ V+ KLGMH
Sbjct: 87 LNDDFEKEIFLDVACFFIGMDRNDVTLILNGCELFGEIGISILVERSLVTVDGKNKLGMH 266
Query: 483 ALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCFD 542
LLRDMGREI+RE SPEE E+ RLW H+DV++VL++ TGTK I+GL +KL + N CF
Sbjct: 267 DLLRDMGREIIREKSPEEIEERCRLWFHDDVLHVLSEQTGTKTIKGLALKLQRANEKCFS 446
Query: 543 TIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKH 602
T AF+KM RLRLLQL V+++GD+K + LRWLSW F + P NFY NL ++EL++
Sbjct: 447 TKAFKKMKRLRLLQLAGVKLVGDFKYISRSLRWLSWNRFSSTHIPTNFYIXNLXSIELEN 626
Query: 603 SNL 605
SN+
Sbjct: 627 SNI 635
Score = 38.1 bits (87), Expect(2) = 5e-62
Identities = 14/25 (56%), Positives = 22/25 (88%)
Frame = +1
Query: 396 EWRSVLSKLEKIPNDQIHEILKISF 420
EW+S L KL++IPN+Q+H+ L+IS+
Sbjct: 4 EWKSALDKLKRIPNNQVHKKLRISY 78
>TC92944 similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (12%)
Length = 673
Score = 209 bits (532), Expect(2) = 1e-58
Identities = 102/164 (62%), Positives = 125/164 (76%)
Frame = +3
Query: 536 TNRVCFDTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNL 595
T + +T AFEKM +LR LQL +Q+ GDYK +HLRWLSW GFPLKY P +F+Q L
Sbjct: 78 TQQPYLETKAFEKMDKLRFLQLVGIQLNGDYKYLSRHLRWLSWHGFPLKYIPADFHQDTL 257
Query: 596 VAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEV 655
VA+ LK+SNL +VW+K Q + LKILNLSHS NL+ TPDFSKLPNLEKLI+KDC SL V
Sbjct: 258 VAVVLKYSNLERVWRKSQFLVKLKILNLSHSHNLRHTPDFSKLPNLEKLILKDCPSLSSV 437
Query: 656 HPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSK 699
+IG LK +L++NLKDCT L LPR IY+L ++TLILSGC+K
Sbjct: 438 SSNIGHLKKILLINLKDCTGLRELPRSIYKLDSLKTLILSGCTK 569
Score = 37.0 bits (84), Expect(2) = 1e-58
Identities = 18/23 (78%), Positives = 21/23 (91%)
Frame = +1
Query: 699 KIDKLEEDIVQMESLTTLMAANT 721
KIDKLEEDI QM+SLTTL+A +T
Sbjct: 568 KIDKLEEDIEQMKSLTTLVADDT 636
>BQ148280 similar to GP|7107246|gb| unknown {Cicer arietinum}, partial (98%)
Length = 668
Score = 218 bits (556), Expect = 6e-57
Identities = 112/222 (50%), Positives = 164/222 (73%), Gaps = 2/222 (0%)
Frame = +1
Query: 216 GKSTIAKVIYNDLCYEFENQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQ 275
GK+TIAK YN + ++F+ +SFL N+RE WE D G++ LQ++ LSDI KT +IK+ ++E
Sbjct: 1 GKTTIAKAAYNKIRHDFDAKSFLLNVREDWEHDNGQVSLQQRLLSDIYKTTEIKIRTLES 180
Query: 276 GKTMIKQQLRAKRILAVLDDVSELEQFDALCQRNS-VGPGSIIIITTRDLRVLNILEVDF 334
GK ++K++L+ K+I VLDDV++ +Q +ALC + G GS IIITTRD +L+ L+V +
Sbjct: 181 GKMILKERLQKKKIFLVLDDVNKEDQLNALCGSHEWFGEGSRIIITTRDDDLLSRLKVHY 360
Query: 335 IYEAEELNASESLELFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLF-KRK 393
+Y +E++ +ESLELF HAF++ P + F LS DVV Y GG+PLAL+V+GS+L +R+
Sbjct: 361 VYRMKEMDDNESLELFSWHAFKQPNPIKGFGNLSTDVVKYSGGLPLALQVIGSFLLTRRR 540
Query: 394 KQEWRSVLSKLEKIPNDQIHEILKISFDGLKDRMEKNIFLDV 435
K+EW S+L KL+ IPND++ E L++SFDGL D K IFLD+
Sbjct: 541 KKEWTSLLEKLKLIPNDKVLEKLQLSFDGLSDDDMKEIFLDI 666
>BF643292 weakly similar to GP|22037340|gb| disease resistance-like protein
GS3-4 {Glycine max}, partial (45%)
Length = 576
Score = 205 bits (522), Expect(2) = 7e-55
Identities = 101/161 (62%), Positives = 124/161 (76%)
Frame = +1
Query: 348 ELFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKI 407
E F HAF++A P++DF +S +VV Y G +PLALEVLGSYLF R+ EW VL KL++I
Sbjct: 1 EFFSWHAFKQARPSKDFSEISTNVVQYSGRLPLALEVLGSYLFDREVTEWICVLEKLKRI 180
Query: 408 PNDQIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIE 467
PNDQ+H+ LKIS+DGL D EK+IFLD+ CFFIG DR V ILNG G A+IGI+VL+E
Sbjct: 181 PNDQVHQKLKISYDGLNDDTEKSIFLDIACFFIGMDRNDVIHILNGSGFFAEIGISVLVE 360
Query: 468 RSLIKVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLW 508
RSL+ V+ KLGMH LLRDMGREI+RE SP EPE+ +RLW
Sbjct: 361 RSLVTVDDKNKLGMHDLLRDMGREIIREKSPMEPEERSRLW 483
Score = 28.1 bits (61), Expect(2) = 7e-55
Identities = 11/18 (61%), Positives = 16/18 (88%)
Frame = +3
Query: 517 LADYTGTKAIEGLVMKLP 534
L+++TGTKA EGL +K+P
Sbjct: 510 LSEHTGTKAGEGLTLKMP 563
>TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll resistance
gene analog protein {Medicago edgeworthii}, partial
(60%)
Length = 942
Score = 211 bits (536), Expect = 1e-54
Identities = 120/305 (39%), Positives = 190/305 (61%), Gaps = 3/305 (0%)
Frame = +1
Query: 207 VGIWGMGGIGKSTIAKVIYNDLC--YEFENQSFLANIREVWEKDRGRIDLQEQFLSDILK 264
VGIW M GIGK+T+A V+Y+ + Y+F+ F+ ++ +++ +D G I +Q++ L +K
Sbjct: 1 VGIWRMDGIGKTTLANVLYDTISHQYQFDACCFVEDVSKIY-RDGGAIAVQKRILDQTIK 177
Query: 265 TRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFDAL-CQRNSVGPGSIIIITTRD 323
+ ++ S + +I +L ++L VLD+V + Q L S+ GS IIITTRD
Sbjct: 178 EKNLEGYSPSEISGIISNRLYKLKLLLVLDNVDQSVQLQELHINPISLCAGSRIIITTRD 357
Query: 324 LRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALE 383
+L E D +YEAE LN +++ EL C+ AF+ + D+ L +V+ Y G+PLA+
Sbjct: 358 KHILIEYEADIVYEAELLNDNDAHELLCRKAFKSDYSSSDYEELIPEVLKYAQGLPLAIR 537
Query: 384 VLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDGLKDRMEKNIFLDVCCFFIGKD 443
V+GS+L+KRK +WR+ L + P+ I ++L+ SF+GL+ R EK IFL V CFF G+
Sbjct: 538 VMGSFLYKRKTAQWRAALEGWQNNPDSGIMKVLRSSFEGLELR-EKEIFLHVACFFDGER 714
Query: 444 RAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGMHALLRDMGREIVRESSPEEPEK 503
YV +IL+ CGL +IGI +E+SLI + +N++ MH +L+++G VRE+ P+EP
Sbjct: 715 EDYVRRILHACGLQPNIGIPTSVEKSLITI-RNQENHMHEMLQELGETNVRETHPDEP-- 885
Query: 504 HTRLW 508
RLW
Sbjct: 886 --RLW 894
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 209 bits (532), Expect = 4e-54
Identities = 131/338 (38%), Positives = 200/338 (58%), Gaps = 11/338 (3%)
Frame = +2
Query: 254 LQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFDALCQRNS-VG 312
LQE+ L L+ KIK+ V +G IK++L K+ L +LDDV ++EQ AL G
Sbjct: 2 LQEELLLKALQL-KIKLGGVSEGIPYIKERLHTKKTLLILDDVDDMEQLHALAGGPDWFG 178
Query: 313 PGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRK-AIPTQDFLILSRDV 371
GS +IITTRD +L ++ +E EEL +E+LEL AF+ +P+ +L+R
Sbjct: 179 RGSRVIITTRDKHLLRSHGIESTHEVEELYGTEALELLRWMAFKNNKVPSIYEDVLNR-A 355
Query: 372 VAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDGLKDRMEKNI 431
V+Y G+PL LE++GS LF + +EW+ L EKIPN +IH+ILK+S+D L++ ++++
Sbjct: 356 VSYASGLPLVLEIVGSNLFGKTIEEWKGTLDGYEKIPNKKIHQILKVSYDALEEE-QQSV 532
Query: 432 FLDVCCFFIG---KDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNK-----KLGMHA 483
FLD+ C F G ++ Y+ + G + + VL E+SL+K+ +L +H
Sbjct: 533 FLDIACCFKGCGWEEFEYILRAHYGHRITHH--LVVLAEKSLVKITHPHYGSIYELTLHD 706
Query: 484 LLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCFDT 543
L+++MG+E+VR+ SP+EP + +RLWC +D+VNVL + TGT IE + M P V
Sbjct: 707 LIKEMGKEVVRQESPKEPGERSRLWCEDDIVNVLKENTGTSKIEMIYMNFPSEEFVIDKK 886
Query: 544 -IAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQG 580
AF+KM RL+ L ++NV K P LR L +G
Sbjct: 887 GKAFKKMTRLKTLIIENVHFSKGLKYLPSSLRVLKLRG 1000
>BQ148669 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (63%)
Length = 608
Score = 119 bits (297), Expect(2) = 1e-50
Identities = 58/96 (60%), Positives = 73/96 (75%)
Frame = +2
Query: 364 FLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDGL 423
F +S +VV YCGG+PLALEVLGSYLF R+ +W +L KL++IPNDQ+ + LKIS+DGL
Sbjct: 29 FAQISINVV*YCGGLPLALEVLGSYLFDRQVTKWECLLEKLKRIPNDQVQKKLKISYDGL 208
Query: 424 KDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNAD 459
D E++IFLD+ FFIG DR V ILNGCGL A+
Sbjct: 209 NDDTERDIFLDIAFFFIGMDRNDVMDILNGCGLFAE 316
Score = 100 bits (249), Expect(2) = 1e-50
Identities = 53/96 (55%), Positives = 67/96 (69%)
Frame = +3
Query: 461 GITVLIERSLIKVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADY 520
GI+VL+ERSL+ ++ KLGMH LLRDMGREI+R+ SP++ EK +RLW HEDV +V
Sbjct: 321 GISVLVERSLVTIDDKNKLGMHDLLRDMGREIIRQKSPKKLEKRSRLWFHEDVHDVFVIT 500
Query: 521 TGTKAIEGLVMKLPKTNRVCFDTIAFEKMIRLRLLQ 556
+ GLV+KL N CF T AFE M +LRLLQ
Sbjct: 501 KWCTSC*GLVLKL-AANAKCFSTNAFENMKKLRLLQ 605
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,297,762
Number of Sequences: 36976
Number of extensions: 378855
Number of successful extensions: 2517
Number of sequences better than 10.0: 317
Number of HSP's better than 10.0 without gapping: 2297
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2359
length of query: 882
length of database: 9,014,727
effective HSP length: 105
effective length of query: 777
effective length of database: 5,132,247
effective search space: 3987755919
effective search space used: 3987755919
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC145513.6


