

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145513.1 - phase: 0 /pseudo
(902 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
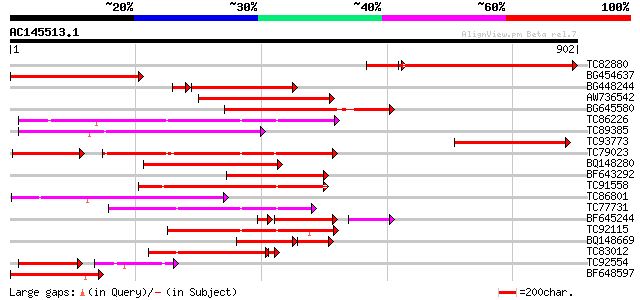
Score E
Sequences producing significant alignments: (bits) Value
TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein ... 506 e-143
BG454637 weakly similar to GP|23477203|emb TIR-NBS disease resis... 429 e-120
BG448244 weakly similar to PIR|T08833|T088 disease resistance pr... 330 e-101
AW736542 similar to PIR|T07656|T076 probable resistance protein ... 348 5e-96
BG645580 similar to PIR|T07656|T076 probable resistance protein ... 295 7e-80
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 290 2e-78
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 250 2e-66
TC93773 weakly similar to GP|9858483|gb|AAG01054.1| resistance p... 247 1e-65
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 233 2e-61
BQ148280 similar to GP|7107246|gb| unknown {Cicer arietinum}, pa... 223 3e-58
BF643292 weakly similar to GP|22037340|gb| disease resistance-li... 207 1e-53
TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll... 206 3e-53
TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - ... 204 2e-52
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 190 2e-48
BF645244 weakly similar to PIR|T07656|T076 probable resistance p... 137 7e-48
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 187 2e-47
BQ148669 similar to PIR|T07656|T076 probable resistance protein ... 120 8e-46
TC83012 weakly similar to GP|7107242|gb|AAF36335.1| unknown {Cic... 174 1e-45
TC92554 similar to GP|9758876|dbj|BAB09430.1 disease resistance ... 115 9e-45
BF648597 weakly similar to GP|12003378|gb Avr9/Cf-9 rapidly elic... 167 1e-41
>TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (46%)
Length = 1238
Score = 506 bits (1304), Expect = e-143
Identities = 271/284 (95%), Positives = 273/284 (95%)
Frame = +3
Query: 619 NTVISHKFGRSLS**RD*RS*ISVILST*KEPLTFQSYQT*KS*L*RIVKVCWRYTRPLE 678
N + H F S S**RD*RS*ISVILST*KEP TFQ+YQT*KS*L*RIVKVCWRYTRPLE
Sbjct: 264 NFYLKHLF--SCS**RD*RS*ISVILST*KEPPTFQNYQT*KS*L*RIVKVCWRYTRPLE 437
Query: 679 ISRIFFC*I*RTVRVLAISQERSIN*EQWKLSSFLVVQRLTNWKKI*CKWNP*QL*WQQI 738
IS IFF *I*RTVRVLAISQERSIN*EQ KLSSFLVVQRLTNWKKI*CKWNP*QL*WQQI
Sbjct: 438 ISTIFF**I*RTVRVLAISQERSIN*EQ*KLSSFLVVQRLTNWKKI*CKWNP*QL*WQQI 617
Query: 739 LESNNHHFR**DQKALDIYLYVDMKDYHIMFFLLSLGLGCHQQ*IL*HIFPLLVACQSLL 798
LESNNHHFR**DQKALDIYLYVDMKDYHIMFFLLSLGLGCHQQ*IL*HIFPLLVACQSLL
Sbjct: 618 LESNNHHFR**DQKALDIYLYVDMKDYHIMFFLLSLGLGCHQQ*IL*HIFPLLVACQSLL 797
Query: 799 LPWI*RAIIWL*SINHKYLAAVQNLEVFRYNVTQRFN*NKNSEDFLMIYMMPVLLNWEYH 858
LPWI*RAIIWL*SINHKYLAAVQNLEVFRYNVTQRFN*NKNSEDFLMIYMMPVLLNWEYH
Sbjct: 798 LPWI*RAIIWL*SINHKYLAAVQNLEVFRYNVTQRFN*NKNSEDFLMIYMMPVLLNWEYH 977
Query: 859 MHHTFRIIP*DLF*LEWEIAT*SSIFLGRAYHRFLLNLLT*YFI 902
MHHTFRIIP*DLF*LEWEIAT*SSIFLGRAYHRFLLNLLT*YFI
Sbjct: 978 MHHTFRIIP*DLF*LEWEIAT*SSIFLGRAYHRFLLNLLT*YFI 1109
Score = 105 bits (263), Expect = 6e-23
Identities = 54/63 (85%), Positives = 58/63 (91%)
Frame = +1
Query: 568 RD*DCCNLIMYKSSEITSVFQST*DGFLGKDFH*NTHQKTFIRKM*LLWT*NTVISHKFG 627
R+ DCCNLIMYKSSEI SVF+S+*DGFLGKDF *NT QKTFIRKM*LLWT*NTVI H+FG
Sbjct: 1 RNCDCCNLIMYKSSEIMSVFRSS*DGFLGKDFL*NTCQKTFIRKM*LLWT*NTVILHRFG 180
Query: 628 RSL 630
RSL
Sbjct: 181 RSL 189
>BG454637 weakly similar to GP|23477203|emb TIR-NBS disease resistance
protein {Populus balsamifera subsp. trichocarpa},
partial (16%)
Length = 659
Score = 429 bits (1102), Expect = e-120
Identities = 212/212 (100%), Positives = 212/212 (100%)
Frame = +3
Query: 1 MSSSSSRSRPQWIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPE 60
MSSSSSRSRPQWIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPE
Sbjct: 9 MSSSSSRSRPQWIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPE 188
Query: 61 LVRAIQGSQIAIVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHSP 120
LVRAIQGSQIAIVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHSP
Sbjct: 189 LVRAIQGSQIAIVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHSP 368
Query: 121 VILVDELDQIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPNF 180
VILVDELDQIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPNF
Sbjct: 369 VILVDELDQIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPNF 548
Query: 181 QVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGG 212
QVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGG
Sbjct: 549 QVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGG 644
>BG448244 weakly similar to PIR|T08833|T088 disease resistance protein
homolog RLG6 - soybean (fragment), partial (34%)
Length = 599
Score = 330 bits (846), Expect(2) = e-101
Identities = 165/169 (97%), Positives = 165/169 (97%)
Frame = +2
Query: 289 AVLDDVSELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELF 348
AVLDDVSELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELF
Sbjct: 92 AVLDDVSELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELF 271
Query: 349 CGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPND 408
CGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPND
Sbjct: 272 CGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPND 451
Query: 409 QIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHAD 457
QIHEKLKI FNGLSDRME DIFLDVCCFFIGKDR YVTKILNG GLHAD
Sbjct: 452 QIHEKLKIXFNGLSDRMEXDIFLDVCCFFIGKDRXYVTKILNGXGLHAD 598
Score = 56.6 bits (135), Expect(2) = e-101
Identities = 29/29 (100%), Positives = 29/29 (100%)
Frame = +1
Query: 259 DILKTRKIKVLSVEQGKAMIKQRLRSKRI 287
DILKTRKIKVLSVEQGKAMIKQRLRSKRI
Sbjct: 1 DILKTRKIKVLSVEQGKAMIKQRLRSKRI 87
>AW736542 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (83%)
Length = 656
Score = 348 bits (893), Expect = 5e-96
Identities = 174/218 (79%), Positives = 192/218 (87%), Gaps = 1/218 (0%)
Frame = +2
Query: 301 NALCEG-NSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPT 359
NALC N +GPGS+IIITTRD R+L+IL VDFIYEAEGLN ES LF HAF++ P+
Sbjct: 2 NALCGNRNGIGPGSIIIITTRDARLLDILGVDFIYEAEGLNVHESRRLFNWHAFKEANPS 181
Query: 360 EDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPNDQIHEKLKISFN 419
E FLILS VV+YCGG+PLALEVLGSYL RRK+EWQSV+SKL+KIPNDQIHEKLKISF+
Sbjct: 182 EAFLILSGDVVSYCGGLPLALEVLGSYLFNRRKREWQSVISKLQKIPNDQIHEKLKISFD 361
Query: 420 GLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGM 479
GL D MEK+IFLDVCCFFIGKDRAYVT+ILNGCGLHADIGI VLIERSL+KVEKN KLGM
Sbjct: 362 GLEDHMEKNIFLDVCCFFIGKDRAYVTEILNGCGLHADIGIEVLIERSLLKVEKNNKLGM 541
Query: 480 HDLLRDMGREIVRESSPEEPEKRTRLWCHEDVVNVLED 517
H LLRDMGREIVRESSPEEPEKRTRLWC EDVV+VL +
Sbjct: 542 HALLRDMGREIVRESSPEEPEKRTRLWCFEDVVDVLAE 655
>BG645580 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (87%)
Length = 814
Score = 295 bits (754), Expect = 7e-80
Identities = 162/272 (59%), Positives = 195/272 (71%), Gaps = 3/272 (1%)
Frame = +1
Query: 343 ESLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKL 402
+ LELF HAFR+ P ++F LSR VVAYCGG+PLALEV+GSYL R KQEW+SVL KL
Sbjct: 1 DPLELFSWHAFRQPSPIKNFSELSRTVVAYCGGLPLALEVIGSYLYGRTKQEWESVLLKL 180
Query: 403 EKIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITV 462
E+IPNDQ+ EKL+IS++GL D M KDIFLD+CCFFIGKDRAYVT+ILNGCGL+ADIGITV
Sbjct: 181 ERIPNDQVQEKLRISYDGLKDDMAKDIFLDICCFFIGKDRAYVTEILNGCGLYADIGITV 360
Query: 463 LIERSLIKVEKNKKLGMHDLLRDMGREIVRESSPEEPEKRTRLWCHEDVVNVLEDHTTTN 522
L+ERSL+K+EKN KLGMHDLLRDMGREIVR+SS + P KR+RLW HEDV +VL +T T
Sbjct: 361 LVERSLVKIEKNNKLGMHDLLRDMGREIVRQSSAKNPGKRSRLWFHEDVHDVLTKNTGT- 537
Query: 523 *DFLFILFFYGLSLCHREQKPSRDWL*SCQRPIEFALIQLLLRK---*RD*DCCNLIMYK 579
+ G+ * CQ P EFA + +L K * +*+ NLI+
Sbjct: 538 ------INVEGVV------------F*VCQEPAEFASVLILSWK*NN*NN*NFYNLIVLT 663
Query: 580 SSEITSVFQST*DGFLGKDFH*NTHQKTFIRK 611
S EI VF + *DG + KD * +Q TFIRK
Sbjct: 664 SLEIMGVFPNN*DGSVCKDLP*TVYQMTFIRK 759
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 290 bits (742), Expect = 2e-78
Identities = 191/530 (36%), Positives = 308/530 (58%), Gaps = 19/530 (3%)
Frame = +3
Query: 14 HDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDEN--LKKGEELGPELVRAIQGSQIA 71
+DVF+ +G DTR F +L AL D GI TF DD++ L++ +++ P + I+ S+I
Sbjct: 90 YDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDKVTPII---IEESRIL 260
Query: 72 IVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHSP---VILVDELD 128
I +FS NY +SS CL+ L I+ C G +V+PVF G+ P+++R H+ L + +
Sbjct: 261 IPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVRHHTGRYGKALAEHEN 440
Query: 129 QIIFGKKR---------ALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPN 179
+ K AL + L + ++ + +++ +IV + + ++ L +
Sbjct: 441 RFQNDTKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGKIVKYISNKISRQSLHVAT 620
Query: 180 FQVGLKPRAEKPIRFLRQNTRK-VCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVAN 238
+ VGL+ R ++ L + V +VGI+G+GG GKST+A+ IYN + +FE F+
Sbjct: 621 YPVGLQSRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLARAIYNFVADQFEGLCFLEQ 800
Query: 239 IREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELE 298
+RE + + QE LLS L+ KIK+ V +G ++IK+RL K+IL +LDDV ++
Sbjct: 801 VRENSASNSLK-RFQEMLLSKTLQL-KIKLADVSEGISIIKERLCRKKILLILDDVDNMK 974
Query: 299 QFNALCEG-NSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRK-V 356
Q NAL G + GPGS +IITTRD +L E++ Y +GLN +E+LEL AF+
Sbjct: 975 QLNALAGGVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNVTEALELLRWMAFKNDK 1154
Query: 357 IPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPNDQIHEKLKI 416
+P+ IL+R VVAY G+P+ +E++GS L + +E ++ L EKIPN +I LK+
Sbjct: 1155VPSSYEKILNR-VVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNKEIQRILKV 1331
Query: 417 SFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHA-DIGITVLIERSLI-KVEKN 474
S++ L + E+ +FLD+ C F G V +IL+ H + + VL+E+ LI E +
Sbjct: 1332SYDSLEEE-EQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHVEVLVEKCLIDHFEYD 1508
Query: 475 KKLGMHDLLRDMGREIVRESSPEEPEKRTRLWCHEDVVNVLEDHTTTN*D 524
+ +H+L+ +MG+E+VR SP EP KR+RLW +D+ VLE++T + D
Sbjct: 1509SHVSLHNLIENMGKELVRLESPFEPGKRSRLWFEKDIFEVLEENTVSKID 1658
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 250 bits (638), Expect = 2e-66
Identities = 143/406 (35%), Positives = 232/406 (56%), Gaps = 12/406 (2%)
Frame = +3
Query: 14 HDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPELVRAIQGSQIAIV 73
+DVF+ FRG+DTR F + L+AAL GI F DD NL KGE +GPEL+R I+GSQ+ +
Sbjct: 69 YDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRTIEGSQVFVA 248
Query: 74 VFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHSPVILVD-------- 125
V S+NY +S+WCL ELE+I +C +G+ V+P+F G+ PS +++ S + D
Sbjct: 249 VLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYWDDFAKHEQRF 428
Query: 126 -ELDQIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPNFQVGL 184
+ + + AL V + GWD+ + QS V++IV +L L K + VG+
Sbjct: 429 KQDPHKVSRWREALNQVGSIAGWDLRD-KQQSVEVEKIVQTILNILKCKSSFVSKDLVGI 605
Query: 185 KPRAEK-PIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVW 243
R E + L + V ++GIWGMGG GK+T+A +Y +C+ F+ F+ ++ +++
Sbjct: 606 NSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFIDDVSKIF 785
Query: 244 EKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNAL 303
G ID Q+Q+L L ++ + +I+ RL ++ L +LD+V ++EQ +
Sbjct: 786 RLHDGPIDAQKQILHQTLGIEHHQICNHYSATDLIRHRLSREKTLLILDNVDQVEQLERI 965
Query: 304 -CEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFR-KVIPTED 361
+G GS I+I +RD +L +VD Y+ L+ +ES +LF AF+ + I ++
Sbjct: 966 GVHREWLGAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAFKLEKIIMKN 1145
Query: 362 FLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPN 407
+ L+ ++ Y G+PL + VLGS+L R EW+S L++L + PN
Sbjct: 1146YQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRPN 1283
>TC93773 weakly similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (31%)
Length = 856
Score = 247 bits (631), Expect = 1e-65
Identities = 135/184 (73%), Positives = 145/184 (78%)
Frame = +3
Query: 708 KLSSFLVVQRLTNWKKI*CKWNP*QL*WQQILESNNHHFR**DQKALDIYLYVDMKDYHI 767
K S FLVVQRL NWKKI*CK N *Q * QQ L SN FR +QK LDIY YV+MK YH+
Sbjct: 18 KTSFFLVVQRLKNWKKI*CK*NH*QP*LQQKLVSNKFLFRLLNQKTLDIYPYVNMKVYHV 197
Query: 768 MFFLLSLGLGCHQQ*IL*HIFPLLVACQSLLLPWI*RAIIWL*SINHKYLAAVQNLEVFR 827
MFFLLS GLGCHQ *I *HIFP LVACQ LL W+* IW+ SINH+YLAA+QNLEVF+
Sbjct: 198 MFFLLSFGLGCHQT*IP*HIFPPLVACQCLLFAWM*IVEIWVWSINHQYLAAIQNLEVFQ 377
Query: 828 YNVTQRFN*NKNSEDFLMIYMMPVLLNWEYHMHHTFRIIP*DLF*LEWEIAT*SSIFLGR 887
YNVTQRFN*NKNSEDFLMIYMM VLLN E+HM H F I P*DLF LE E+AT*SSI GR
Sbjct: 378 YNVTQRFN*NKNSEDFLMIYMMQVLLNLEHHMDHKFWIFP*DLFCLESEVAT*SSILFGR 557
Query: 888 AYHR 891
AY R
Sbjct: 558 AYQR 569
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 233 bits (594), Expect = 2e-61
Identities = 149/380 (39%), Positives = 235/380 (61%), Gaps = 6/380 (1%)
Frame = +3
Query: 148 DMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPNFQVGLKPRAEKPIRFLRQNTR-KVCLVG 206
DM+ Y + + + +IV + + ++ L + + VGL+ R + L + + +V +VG
Sbjct: 678 DMNRY--EYEFIGKIVEDISNRISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVG 851
Query: 207 IWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRK- 265
++G GGIGKST+AK IYN + +FE F+ N+R D + LQE+LL LKT +
Sbjct: 852 LYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLK-HLQEKLL---LKTVRL 1019
Query: 266 -IKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNALCEG-NSVGPGSVIIITTRDLR 323
IK+ V QG +IKQRL K+IL +LDDV +L+Q AL G + GPGS +IITTR+
Sbjct: 1020 DIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKH 1199
Query: 324 VLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVL 383
+L I ++ + EGLNA+E+LEL AF++ +P+ IL+R + Y G+PLA+ ++
Sbjct: 1200 LLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNR-ALTYASGLPLAIVII 1376
Query: 384 GSYLLKRRKQEWQSVLSKLEKIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRA 443
GS L+ R Q+ S L E+IPN +I LK+S++ L ++ E+ +FLD+ C F G
Sbjct: 1377 GSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDSL-EKEEQSVFLDIACCFKGCKWP 1553
Query: 444 YVTKILNGCGLHADIG-ITVLIERSLIK-VEKNKKLGMHDLLRDMGREIVRESSPEEPEK 501
V +IL+ H + + VL E+SL+ ++ + + +HDL+ DMG+E+VR+ SP+EP +
Sbjct: 1554 EVKEILHAHYGHCIVHHVAVLAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEPGE 1733
Query: 502 RTRLWCHEDVVNVLEDHTTT 521
R+RLW D+V+VL+ +T T
Sbjct: 1734 RSRLWFERDIVHVLKKNTGT 1793
Score = 114 bits (286), Expect = 1e-25
Identities = 54/115 (46%), Positives = 81/115 (69%)
Frame = +1
Query: 5 SSRSRPQWIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPELVRA 64
+++S + + VF++FRG DTR F +LY ALTD GI TF+DD +L++G+E+ P L A
Sbjct: 67 ATQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNA 246
Query: 65 IQGSQIAIVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHS 119
I+ S+I I VFS+NY +SS+CL+EL I C G +V+PVF G+ P+++R H+
Sbjct: 247 IEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHT 411
>BQ148280 similar to GP|7107246|gb| unknown {Cicer arietinum}, partial (98%)
Length = 668
Score = 223 bits (568), Expect = 3e-58
Identities = 115/222 (51%), Positives = 167/222 (74%), Gaps = 2/222 (0%)
Frame = +1
Query: 214 GKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQ 273
GK+TIAK YN + ++F+ +SF+ N+RE WE D G++ LQ++LLSDI KT +IK+ ++E
Sbjct: 1 GKTTIAKAAYNKIRHDFDAKSFLLNVREDWEHDNGQVSLQQRLLSDIYKTTEIKIRTLES 180
Query: 274 GKAMIKQRLRSKRILAVLDDVSELEQFNALCEGNS-VGPGSVIIITTRDLRVLNILEVDF 332
GK ++K+RL+ K+I VLDDV++ +Q NALC + G GS IIITTRD +L+ L+V +
Sbjct: 181 GKMILKERLQKKKIFLVLDDVNKEDQLNALCGSHEWFGEGSRIIITTRDDDLLSRLKVHY 360
Query: 333 IYEAEGLNASESLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLL-KRR 391
+Y + ++ +ESLELF HAF++ P + F LS VV Y GG+PLAL+V+GS+LL +RR
Sbjct: 361 VYRMKEMDDNESLELFSWHAFKQPNPIKGFGNLSTDVVKYSGGLPLALQVIGSFLLTRRR 540
Query: 392 KQEWQSVLSKLEKIPNDQIHEKLKISFNGLSDRMEKDIFLDV 433
K+EW S+L KL+ IPND++ EKL++SF+GLSD K+IFLD+
Sbjct: 541 KKEWTSLLEKLKLIPNDKVLEKLQLSFDGLSDDDMKEIFLDI 666
>BF643292 weakly similar to GP|22037340|gb| disease resistance-like protein
GS3-4 {Glycine max}, partial (45%)
Length = 576
Score = 207 bits (527), Expect = 1e-53
Identities = 101/161 (62%), Positives = 123/161 (75%)
Frame = +1
Query: 346 ELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKI 405
E F HAF++ P++DF +S VV Y G +PLALEVLGSYL R EW VL KL++I
Sbjct: 1 EFFSWHAFKQARPSKDFSEISTNVVQYSGRLPLALEVLGSYLFDREVTEWICVLEKLKRI 180
Query: 406 PNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVLIE 465
PNDQ+H+KLKIS++GL+D EK IFLD+ CFFIG DR V ILNG G A+IGI+VL+E
Sbjct: 181 PNDQVHQKLKISYDGLNDDTEKSIFLDIACFFIGMDRNDVIHILNGSGFFAEIGISVLVE 360
Query: 466 RSLIKVEKNKKLGMHDLLRDMGREIVRESSPEEPEKRTRLW 506
RSL+ V+ KLGMHDLLRDMGREI+RE SP EPE+R+RLW
Sbjct: 361 RSLVTVDDKNKLGMHDLLRDMGREIIREKSPMEPEERSRLW 483
>TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll resistance
gene analog protein {Medicago edgeworthii}, partial
(60%)
Length = 942
Score = 206 bits (524), Expect = 3e-53
Identities = 120/305 (39%), Positives = 187/305 (60%), Gaps = 3/305 (0%)
Frame = +1
Query: 205 VGIWGMGGIGKSTIAKVIYNDLC--YEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILK 262
VGIW M GIGK+T+A V+Y+ + Y+F+ FV ++ +++ +D G I +Q+++L +K
Sbjct: 1 VGIWRMDGIGKTTLANVLYDTISHQYQFDACCFVEDVSKIY-RDGGAIAVQKRILDQTIK 177
Query: 263 TRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNAL-CEGNSVGPGSVIIITTRD 321
+ ++ S + +I RL ++L VLD+V + Q L S+ GS IIITTRD
Sbjct: 178 EKNLEGYSPSEISGIISNRLYKLKLLLVLDNVDQSVQLQELHINPISLCAGSRIIITTRD 357
Query: 322 LRVLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALE 381
+L E D +YEAE LN +++ EL C AF+ + D+ L V+ Y G+PLA+
Sbjct: 358 KHILIEYEADIVYEAELLNDNDAHELLCRKAFKSDYSSSDYEELIPEVLKYAQGLPLAIR 537
Query: 382 VLGSYLLKRRKQEWQSVLSKLEKIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKD 441
V+GS+L KR+ +W++ L + P+ I + L+ SF GL R EK+IFL V CFF G+
Sbjct: 538 VMGSFLYKRKTAQWRAALEGWQNNPDSGIMKVLRSSFEGLELR-EKEIFLHVACFFDGER 714
Query: 442 RAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGMHDLLRDMGREIVRESSPEEPEK 501
YV +IL+ CGL +IGI +E+SLI + +N++ MH++L+++G VRE+ P+EP
Sbjct: 715 EDYVRRILHACGLQPNIGIPTSVEKSLITI-RNQENHMHEMLQELGETNVRETHPDEP-- 885
Query: 502 RTRLW 506
RLW
Sbjct: 886 --RLW 894
>TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (7%)
Length = 1268
Score = 204 bits (518), Expect = 2e-52
Identities = 128/359 (35%), Positives = 211/359 (58%), Gaps = 13/359 (3%)
Frame = +2
Query: 3 SSSSRSRPQWIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPELV 62
+SSS S Q +DVFI+FRG+DTR F SHL+AAL+ ++T++D ++KG+E+ PEL
Sbjct: 35 ASSSHSAAQKKYDVFISFRGEDTRAGFTSHLHAALSRTYLHTYID-YRIEKGDEVWPELE 211
Query: 63 RAIQGSQIAIVVFSKNYVNSSWCLNELEQIMKCK---ADNGQVVMPVFNGITPSNIRQHS 119
+AI+ S + +VVFS+NY +S+WCLNEL ++M+C+ D+ V+PVF + PS++R+ +
Sbjct: 212 KAIKQSTLFLVVFSENYASSTWCLNELVELMECRNKNEDDNIGVIPVFYHVDPSHVRKQT 391
Query: 120 PVI--------LVDELDQIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLD 171
++ D+++ K AL + L+G+ S Y +S ++++I +L L+
Sbjct: 392 GSYGSALAKHKQENQDDKMMQNWKNALFQAANLSGFHSSTYRTESNMIEDITRALLGKLN 571
Query: 172 KKYLPLPNFQVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFE 231
+Y + L ++ ++ V ++G+WGMGG GK+T+A ++ +++E
Sbjct: 572 HQYRDELTCNLILDENYWAVRSLIKFDSTTVQIIGLWGMGGTGKTTLAAAMFQRFSFKYE 751
Query: 232 DQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVL 291
F+ + EV K G +LLS +L +++ + + AMIK+RLR + VL
Sbjct: 752 GNCFLERVTEV-SKKHGINYTCNKLLSKLL-GEDLRIDTPKVIPAMIKRRLRHMKSFIVL 925
Query: 292 DDV--SELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELF 348
DDV SEL Q G +GPGS++I+TTRD VL +D IYE + +N+ SL+LF
Sbjct: 926 DDVHNSELLQDLIGVRGGWLGPGSIVIVTTRDKHVLISGGIDEIYEVKKMNSQNSLQLF 1102
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 190 bits (482), Expect = 2e-48
Identities = 127/336 (37%), Positives = 197/336 (57%), Gaps = 5/336 (1%)
Frame = +1
Query: 157 KVVKEIVSQVLKNLDKKYLPLPNFQVGLKPRAEKPIRFLRQNTR-KVCLVGIWGMGGIGK 215
K +++IV + N++ +L + + VGL+ R E+ L + +V +VG++G GG+GK
Sbjct: 508 KFIEKIVEDISNNINHVFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGLFGTGGMGK 687
Query: 216 STIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGK 275
ST+AK +YN + +FE F+ N+RE + LQ++LLS I+K K+ V +G
Sbjct: 688 STLAKAVYNFVADQFEGVCFLHNVREN-STHKNLKHLQKKLLSKIVKFDG-KLEDVSEGI 861
Query: 276 AMIKQRLRSKRILAVLDDVSELEQFNALCEG-NSVGPGSVIIITTRDLRVLNILEVDFIY 334
+IK+RL K+IL +LDDV +LEQ AL G + G GS +IITTRD +L + +
Sbjct: 862 PIIKERLSRKKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLACHGITSTH 1041
Query: 335 EAEGLNASESLELFCGHAFRK-VIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQ 393
E LN +E+LEL AF+ +P+ IL+R VV Y G+PLA+ +G L R+ +
Sbjct: 1042AVEELNETEALELLRRMAFKNDKVPSTYEEILNR-VVTYASGLPLAIVTIGDNLFGRKVE 1218
Query: 394 EWQSVLSKLEKIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCG 453
+W+ +L + E IPN I L++S++ L + EK +FLD+ C F G V KIL+
Sbjct: 1219DWKRILDEYENIPNKDIQRILQVSYDALEPK-EKSVFLDIACCFKGCKWTKVKKILHAHY 1395
Query: 454 LHA-DIGITVLIERSLI-KVEKNKKLGMHDLLRDMG 487
H + + VL E+SLI E + ++ +HDL+ DMG
Sbjct: 1396GHCIEHHVGVLAEKSLIGHWEYDTQMTLHDLIEDMG 1503
Score = 38.9 bits (89), Expect = 0.009
Identities = 17/41 (41%), Positives = 26/41 (62%)
Frame = +2
Query: 79 YVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHS 119
Y +SS+CL+EL I+ C +V+PVF + P++IR S
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQS 124
>BF645244 weakly similar to PIR|T07656|T076 probable resistance protein -
soybean (fragment), partial (81%)
Length = 678
Score = 137 bits (346), Expect(3) = 7e-48
Identities = 66/101 (65%), Positives = 80/101 (78%)
Frame = +3
Query: 421 LSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGMH 480
L+D EK+IFLDV CFFIG DR VT ILNGC L +IGI++L+ERSL+ V+ KLGMH
Sbjct: 87 LNDDFEKEIFLDVACFFIGMDRNDVTLILNGCELFGEIGISILVERSLVTVDGKNKLGMH 266
Query: 481 DLLRDMGREIVRESSPEEPEKRTRLWCHEDVVNVLEDHTTT 521
DLLRDMGREI+RE SPEE E+R RLW H+DV++VL + T T
Sbjct: 267 DLLRDMGREIIREKSPEEIEERCRLWFHDDVLHVLSEQTGT 389
Score = 52.4 bits (124), Expect(3) = 7e-48
Identities = 31/73 (42%), Positives = 43/73 (58%)
Frame = +1
Query: 540 EQKPSRDWL*SCQRPIEFALIQLLLRK*RD*DCCNLIMYKSSEITSVFQST*DGFLGKDF 599
EQK R WL*SC+ ++ + R+*RD DC NL+ EI ++F+ *DG LG DF
Sbjct: 385 EQKLLRGWL*SCKELMKNVSVLKHSRR*RDLDCFNLLGSNLLEILNIFREV*DGCLGIDF 564
Query: 600 H*NTHQKTFIRKM 612
+ + TFI K+
Sbjct: 565 LQHIYLXTFIXKI 603
Score = 40.8 bits (94), Expect(3) = 7e-48
Identities = 15/25 (60%), Positives = 23/25 (92%)
Frame = +1
Query: 394 EWQSVLSKLEKIPNDQIHEKLKISF 418
EW+S L KL++IPN+Q+H+KL+IS+
Sbjct: 4 EWKSALDKLKRIPNNQVHKKLRISY 78
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 187 bits (474), Expect = 2e-47
Identities = 112/279 (40%), Positives = 173/279 (61%), Gaps = 8/279 (2%)
Frame = +2
Query: 252 LQEQLLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNALCEGNS-VG 310
LQE+LL L+ KIK+ V +G IK+RL +K+ L +LDDV ++EQ +AL G G
Sbjct: 2 LQEELLLKALQL-KIKLGGVSEGIPYIKERLHTKKTLLILDDVDDMEQLHALAGGPDWFG 178
Query: 311 PGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRK-VIPTEDFLILSRYV 369
GS +IITTRD +L ++ +E E L +E+LEL AF+ +P+ +L+R
Sbjct: 179 RGSRVIITTRDKHLLRSHGIESTHEVEELYGTEALELLRWMAFKNNKVPSIYEDVLNR-A 355
Query: 370 VAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPNDQIHEKLKISFNGLSDRMEKDI 429
V+Y G+PL LE++GS L + +EW+ L EKIPN +IH+ LK+S++ L + ++ +
Sbjct: 356 VSYASGLPLVLEIVGSNLFGKTIEEWKGTLDGYEKIPNKKIHQILKVSYDALEEE-QQSV 532
Query: 430 FLDVCCFFIGKDRAYVTKILNG-CGLHADIGITVLIERSLIKVEKNK-----KLGMHDLL 483
FLD+ C F G IL G + VL E+SL+K+ +L +HDL+
Sbjct: 533 FLDIACCFKGCGWEEFEYILRAHYGHRITHHLVVLAEKSLVKITHPHYGSIYELTLHDLI 712
Query: 484 RDMGREIVRESSPEEPEKRTRLWCHEDVVNVLEDHTTTN 522
++MG+E+VR+ SP+EP +R+RLWC +D+VNVL+++T T+
Sbjct: 713 KEMGKEVVRQESPKEPGERSRLWCEDDIVNVLKENTGTS 829
>BQ148669 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (63%)
Length = 608
Score = 120 bits (301), Expect(2) = 8e-46
Identities = 58/96 (60%), Positives = 74/96 (76%)
Frame = +2
Query: 362 FLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPNDQIHEKLKISFNGL 421
F +S VV YCGG+PLALEVLGSYL R+ +W+ +L KL++IPNDQ+ +KLKIS++GL
Sbjct: 29 FAQISINVV*YCGGLPLALEVLGSYLFDRQVTKWECLLEKLKRIPNDQVQKKLKISYDGL 208
Query: 422 SDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHAD 457
+D E+DIFLD+ FFIG DR V ILNGCGL A+
Sbjct: 209 NDDTERDIFLDIAFFFIGMDRNDVMDILNGCGLFAE 316
Score = 82.8 bits (203), Expect(2) = 8e-46
Identities = 37/56 (66%), Positives = 48/56 (85%)
Frame = +3
Query: 459 GITVLIERSLIKVEKNKKLGMHDLLRDMGREIVRESSPEEPEKRTRLWCHEDVVNV 514
GI+VL+ERSL+ ++ KLGMHDLLRDMGREI+R+ SP++ EKR+RLW HEDV +V
Sbjct: 321 GISVLVERSLVTIDDKNKLGMHDLLRDMGREIIRQKSPKKLEKRSRLWFHEDVHDV 488
>TC83012 weakly similar to GP|7107242|gb|AAF36335.1| unknown {Cicer
arietinum}, partial (88%)
Length = 654
Score = 174 bits (441), Expect(2) = 1e-45
Identities = 95/198 (47%), Positives = 131/198 (65%), Gaps = 4/198 (2%)
Frame = +1
Query: 222 IYNDLCYEFEDQSFVANIREVWEKDRG---RIDLQEQLLSDILKTRKIKVLSVEQGKAMI 278
IYN + +F +SF+ NIRE E+D I LQ+QLLSD+LKT++ K+ ++ G I
Sbjct: 4 IYNQIHRKFVYRSFIENIRETCERDSKGGWHICLQQQLLSDLLKTKE-KIHNIASGTIAI 180
Query: 279 KQRLRSKRILAVLDDVSELEQFNALCEGNS-VGPGSVIIITTRDLRVLNILEVDFIYEAE 337
K+ L +K++L VLDDV+++EQ AL E G GSV+I+T+RD +L L+VD +Y
Sbjct: 181 KKMLSAKKVLIVLDDVTKVEQVKALYESRKWFGAGSVLIVTSRDAHILKSLQVDHVYPVN 360
Query: 338 GLNASESLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQS 397
++ ESLELF HAFR+ P DF LS V+ YCGG+PLA EV+GSYL R ++EW S
Sbjct: 361 EMDQKESLELFSWHAFRQASPRADFSELSSSVIKYCGGLPLAAEVIGSYLYGRTREEWTS 540
Query: 398 VLSKLEKIPNDQIHEKLK 415
VLSKLE IP+ + + K
Sbjct: 541 VLSKLEIIPDHHVQRETK 594
Score = 28.5 bits (62), Expect(2) = 1e-45
Identities = 12/18 (66%), Positives = 16/18 (88%)
Frame = +2
Query: 412 EKLKISFNGLSDRMEKDI 429
EKL+IS++GLSD +KDI
Sbjct: 584 EKLRISYDGLSDGKQKDI 637
>TC92554 similar to GP|9758876|dbj|BAB09430.1 disease resistance protein
{Arabidopsis thaliana}, partial (7%)
Length = 1043
Score = 115 bits (288), Expect(2) = 9e-45
Identities = 51/103 (49%), Positives = 76/103 (73%)
Frame = +1
Query: 14 HDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPELVRAIQGSQIAIV 73
H VF+NFRG+DTR F SHLYAAL I+TF+DDE +++G+ + P L+ AI+ S+I ++
Sbjct: 235 HHVFLNFRGEDTRYNFTSHLYAALCGKKIHTFMDDEEIERGDNISPTLLSAIESSKICVI 414
Query: 74 VFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIR 116
+FS++Y +SSWCL+EL +I++C VV+PVF I S++R
Sbjct: 415 IFSQDYASSSWCLDELVKIIECSEKKKLVVIPVFYHIDASHVR 543
Score = 84.3 bits (207), Expect(2) = 9e-45
Identities = 48/138 (34%), Positives = 75/138 (53%), Gaps = 4/138 (2%)
Frame = +2
Query: 135 KRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPNFQ----VGLKPRAEK 190
+ AL + L GW +++ ++KEIV +L DK P+ Q VG+
Sbjct: 626 RTALHKAANLAGWVSEKNRSEAVLIKEIVEVIL---DKLKCMSPHVQNKGLVGISVHVAH 796
Query: 191 PIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRI 250
L + V ++GIWGMGGIGK+TIA ++ + Y++E F AN+RE W RI
Sbjct: 797 VESMLCSGSADVHIIGIWGMGGIGKTTIADAVFTKVSYQYERYYFAANVRETW---GNRI 967
Query: 251 DLQEQLLSDILKTRKIKV 268
LQ ++L+D+L+ + IK+
Sbjct: 968 KLQNEVLADVLEDQSIKI 1021
>BF648597 weakly similar to GP|12003378|gb Avr9/Cf-9 rapidly elicited protein
4 {Nicotiana tabacum}, partial (21%)
Length = 630
Score = 167 bits (424), Expect = 1e-41
Identities = 83/160 (51%), Positives = 115/160 (71%), Gaps = 13/160 (8%)
Frame = +1
Query: 2 SSSSSRSRPQWIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPEL 61
SSSSS + PQ++HDVFINFRG+D R+TFVSHLYA L++AGINTFLD+E L+KGE++G EL
Sbjct: 148 SSSSSSTNPQYLHDVFINFRGEDVRRTFVSHLYAVLSNAGINTFLDNEKLEKGEDIGHEL 327
Query: 62 VRAIQGSQIAIVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQH--- 118
++AI S+I+I+VFSKNY SSWCLNELE+IM+C+ +G VV+PVF + PS +R
Sbjct: 328 LQAISVSRISIIVFSKNYTESSWCLNELEKIMECRXLHGHVVLPVFYDVDPSXVRHQKGD 507
Query: 119 ----------SPVILVDELDQIIFGKKRALRDVSYLTGWD 148
S I+ + + + + ++ L + S L+GWD
Sbjct: 508 FGKALEVAAKSRYIIEEVMGKELGKWRKVLTEASNLSGWD 627
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.345 0.153 0.504
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,817,717
Number of Sequences: 36976
Number of extensions: 415790
Number of successful extensions: 3959
Number of sequences better than 10.0: 225
Number of HSP's better than 10.0 without gapping: 1670
Number of HSP's successfully gapped in prelim test: 159
Number of HSP's that attempted gapping in prelim test: 2100
Number of HSP's gapped (non-prelim): 1930
length of query: 902
length of database: 9,014,727
effective HSP length: 105
effective length of query: 797
effective length of database: 5,132,247
effective search space: 4090400859
effective search space used: 4090400859
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC145513.1


