

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.9 - phase: 0
(918 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
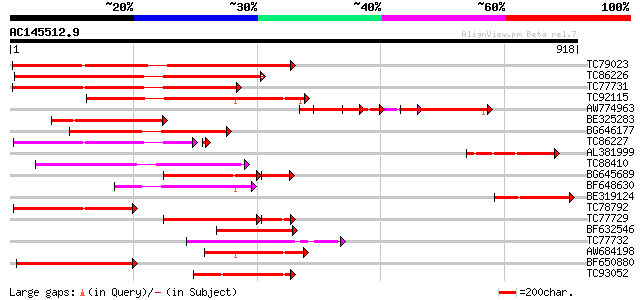
Score E
Sequences producing significant alignments: (bits) Value
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 377 e-104
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 362 e-100
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 300 2e-81
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 300 2e-81
AW774963 weakly similar to GP|22037377|gb| disease resistance-li... 283 2e-76
BE325283 weakly similar to GP|13897750|gb resistance-gene protei... 260 1e-69
BG646177 weakly similar to GP|18033111|g functional candidate re... 221 1e-57
TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 217 4e-57
AL381999 similar to GP|18033111|gb functional candidate resistan... 207 1e-53
TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative res... 206 3e-53
BG645689 similar to GP|18033111|g functional candidate resistanc... 176 3e-51
BF648630 weakly similar to GP|18033111|g functional candidate re... 187 2e-47
BE319124 homologue to GP|18033111|gb functional candidate resist... 178 7e-45
TC78792 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 177 2e-44
TC77729 weakly similar to GP|22037342|gb|AAM90012.1 disease resi... 177 2e-44
BF632546 similar to GP|18033111|gb functional candidate resistan... 174 1e-43
TC77732 similar to GP|9965107|gb|AAG09953.1| resistance protein ... 173 2e-43
AW684198 weakly similar to GP|18033111|g functional candidate re... 166 5e-41
BF650880 weakly similar to GP|9858476|gb| resistance protein LM1... 165 6e-41
TC93052 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 159 5e-39
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 377 bits (967), Expect = e-104
Identities = 208/463 (44%), Positives = 293/463 (62%), Gaps = 5/463 (1%)
Frame = +3
Query: 5 LLLDRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKV 64
+LLD + Y +EFI KIVE++ I L + VG++ + QHV L+ SDD +H V
Sbjct: 669 ILLDMNRYEYEFIGKIVEDISNRISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMV 848
Query: 65 GILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQ-KILLSHIFGEKN 123
G+ G GGIGK+TLA +YN I QFE CFLE VR NS + L +LQ K+LL + +
Sbjct: 849 GLYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTV--RLD 1022
Query: 124 TEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNL 183
++ V +GI I+K+RL KK+LL+LDDVDK +QL+A+AG +WF GSRVIITTR K+L
Sbjct: 1023 IKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHL 1202
Query: 184 LISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKD 243
L G+E + V+GLN +A +L+ W K + ++D+L
Sbjct: 1203 LKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDIL------------------- 1325
Query: 244 LKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQ 303
R + YASG PLA+ +IGS+ ++++ LD Y+ +P+ IQ L+
Sbjct: 1326 -----------NRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILK 1472
Query: 304 LSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLI-KINES 362
+SYDSL++EE+ VFLDIACCFKG K V+ ILHAH+GH + + VL EK L+ +
Sbjct: 1473 VSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMDHLKYD 1652
Query: 363 GNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHF---DG 419
VTLHDL+EDMGKE+VRQE+P++PG+RSRLWF DI+ VL++NTGT KI+MI+
Sbjct: 1653 SYVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDIVHVLKKNTGTRKIKMINMKFPSM 1832
Query: 420 VIEVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLE 462
++ W+G AF+KM NLKT I N S++ ++LP+SLRV++
Sbjct: 1833 ESDIDWNGNAFEKMTNLKTFITENG-HHSKSLEYLPSSLRVMK 1958
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 362 bits (930), Expect = e-100
Identities = 199/408 (48%), Positives = 261/408 (63%), Gaps = 1/408 (0%)
Frame = +3
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
D GY +E I KIV+ + I +L + VG++ + Q V LL+ G DD +H VGI
Sbjct: 531 DSHGYEYELIGKIVKYISNKISRQSLHVATYPVGLQSRVQQVKSLLDEGPDDGVHMVGIY 710
Query: 68 GIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEIT 127
GIGG GK+TLA +YN + QFE CFLE+VRENS N L Q++LLS K ++
Sbjct: 711 GIGGSGKSTLARAIYNFVADQFEGLCFLEQVRENSASNSLKRFQEMLLSKTLQLK-IKLA 887
Query: 128 SVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISS 187
V EGISI+K+RL KK+LL+LDDVD +QL A+AG +WF GSRVIITTR K+LL
Sbjct: 888 DVSEGISIIKERLCRKKILLILDDVDNMKQLNALAGGVDWFGPGSRVIITTRDKHLLACH 1067
Query: 188 GVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKND 247
+E+ Y VKGLN +A +L+ W KND P
Sbjct: 1068EIEKTYAVKGLNVTEALELLRWMAFKNDKVP----------------------------- 1160
Query: 248 EGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYD 307
Y +L RVVAYASG P+ +E++GS+ K IE+C + LD Y+++P+ IQ L++SYD
Sbjct: 1161SSYEKILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNKEIQRILKVSYD 1340
Query: 308 SLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINE-SGNVT 366
SL+EEE+ VFLDIACCFKG K V+ ILHAH+GH + + VLVEK LI E +V+
Sbjct: 1341SLEEEEQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHVEVLVEKCLIDHFEYDSHVS 1520
Query: 367 LHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEM 414
LH+L+E+MGKE+VR E+P +PGKRSRLWF +DI +VLEENT SKI+M
Sbjct: 1521LHNLIENMGKELVRLESPFEPGKRSRLWFEKDIFEVLEENT-VSKIDM 1661
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 300 bits (768), Expect = 2e-81
Identities = 170/372 (45%), Positives = 228/372 (60%), Gaps = 1/372 (0%)
Frame = +1
Query: 5 LLLDRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKV 64
+LLD + Y ++FIEKIVE++ NI V L + VG++ + + V LLL++GS+D + V
Sbjct: 478 ILLDTNRYEYKFIEKIVEDISNNINHVFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMV 657
Query: 65 GILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNT 124
G+ G GG+GK+TLA VYN + QFE CFL VRENS L +LQK LLS I + +
Sbjct: 658 GLFGTGGMGKSTLAKAVYNFVADQFEGVCFLHNVRENSTHKNLKHLQKKLLSKIV-KFDG 834
Query: 125 EITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLL 184
++ V EGI I+K+RL KK+LL+LDDVDK EQL+A+AG +WF GSRVIITTR K+LL
Sbjct: 835 KLEDVSEGIPIIKERLSRKKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLL 1014
Query: 185 ISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDL 244
G+ + V+ LN+ +A +L+ KND P
Sbjct: 1015ACHGITSTHAVEELNETEALELLRRMAFKNDKVP-------------------------- 1116
Query: 245 KNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQL 304
Y +L RVV YASG PLA+ IG + + +E LD Y+ +P+ IQ LQ+
Sbjct: 1117---STYEEILNRVVTYASGLPLAIVTIGDNLFGRKVEDWKRILDEYENIPNKDIQRILQV 1287
Query: 305 SYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINE-SG 363
SYD+L+ +EK VFLDIACCFKG K T V+ ILHAH+GH ++ + VL EK LI E
Sbjct: 1288SYDALEPKEKSVFLDIACCFKGCKWTKVKKILHAHYGHCIEHHVGVLAEKSLIGHWEYDT 1467
Query: 364 NVTLHDLVEDMG 375
+TLHDL+EDMG
Sbjct: 1468QMTLHDLIEDMG 1503
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 300 bits (767), Expect = 2e-81
Identities = 172/373 (46%), Positives = 232/373 (62%), Gaps = 12/373 (3%)
Frame = +2
Query: 125 EITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLL 184
++ V EGI +K+RL KK LL+LDDVD EQL A+AG +WF +GSRVIITTR K+LL
Sbjct: 44 KLGGVSEGIPYIKERLHTKKTLLILDDVDDMEQLHALAGGPDWFGRGSRVIITTRDKHLL 223
Query: 185 ISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDL 244
S G+E +EV+ L +A +L+ W KN+ P
Sbjct: 224 RSHGIESTHEVEELYGTEALELLRWMAFKNNKVPSI------------------------ 331
Query: 245 KNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQL 304
Y +VL R V+YASG PL LE++GS+ KTIE+ LD Y+++P+ I L++
Sbjct: 332 -----YEDVLNRAVSYASGLPLVLEIVGSNLFGKTIEEWKGTLDGYEKIPNKKIHQILKV 496
Query: 305 SYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESG- 363
SYD+L+EE++ VFLDIACCFKG E IL AH+GH + + VL EK L+KI
Sbjct: 497 SYDALEEEQQSVFLDIACCFKGCGWEEFEYILRAHYGHRITHHLVVLAEKSLVKITHPHY 676
Query: 364 ----NVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDG 419
+TLHDL+++MGKE+VRQE+P++PG+RSRLW +DI+ VL+ENTGTSKIEMI+ +
Sbjct: 677 GSIYELTLHDLIKEMGKEVVRQESPKEPGERSRLWCEDDIVNVLKENTGTSKIEMIYMNF 856
Query: 420 VIE---VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*E----LDL 472
E + G+AFKKM LKTLI N V FS+ K+LP+SLRVL+ R + E L
Sbjct: 857 PSEEFVIDKKGKAFKKMTRLKTLIIEN-VHFSKGLKYLPSSLRVLKLRGCLSESLISCSL 1033
Query: 473 SYCINLESFSHVV 485
S I + SFS+ +
Sbjct: 1034SKAIEITSFSYCI 1072
>AW774963 weakly similar to GP|22037377|gb| disease resistance-like protein
GS5-1 {Glycine max}, partial (21%)
Length = 531
Score = 283 bits (725), Expect = 2e-76
Identities = 146/176 (82%), Positives = 147/176 (82%), Gaps = 28/176 (15%)
Frame = +2
Query: 634 KTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKS 693
KTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKS
Sbjct: 2 KTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKS 181
Query: 694 IPPLKCDALETLDLSCCYSLQSFSLVADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCT 753
IPPLKCDALETLDLSCCYSLQSFSLVADRLWKLVLDDCKEL EIKVIPPCLRMLSAVNCT
Sbjct: 182 IPPLKCDALETLDLSCCYSLQSFSLVADRLWKLVLDDCKELHEIKVIPPCLRMLSAVNCT 361
Query: 754 SLTSSCTSKLLN----------------------------QELHKAGNTWFCLPRV 781
S+TSSCTSKLLN QELHKAGNTWFCLPRV
Sbjct: 362 SMTSSCTSKLLNQVYDLFCCYL*FILYSF*YS*T*IIYN*QELHKAGNTWFCLPRV 529
Score = 115 bits (287), Expect = 1e-25
Identities = 64/115 (55%), Positives = 78/115 (67%)
Frame = +2
Query: 493 RTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRS 552
+T+S + C L+SIP LKLD LET+DLS C LES PLVVDG LGK+KTL V +CHNL+S
Sbjct: 2 KTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKS 181
Query: 553 IPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSL 607
IPPLK D+LE LD+S C SL+SF V R L KL + K + IP + L
Sbjct: 182 IPPLKCDALETLDLSCCYSLQSFSLVADR-LWKLVLDDCKELHEIKVIPPCLRML 343
Score = 105 bits (261), Expect = 1e-22
Identities = 65/129 (50%), Positives = 76/129 (58%)
Frame = +2
Query: 540 KTLNVESCHNLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMIS 599
KTL+ +SC NLRSIP LKLD LE LD+S C SLES P V FLGKLKTL V +C + S
Sbjct: 2 KTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKS 181
Query: 600 IPTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKLDLLETLD 659
IP L LE LDLS C +L++F LV D L KL K ++ IP L L
Sbjct: 182 IPPLKCDALETLDLSCCYSLQSFSLVADR-LWKLVLDDCKELHEIKVIP----PCLRMLS 346
Query: 660 LSNCVSLES 668
NC S+ S
Sbjct: 347 AVNCTSMTS 373
Score = 92.8 bits (229), Expect = 5e-19
Identities = 52/105 (49%), Positives = 66/105 (62%)
Frame = +2
Query: 470 LDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFP 529
LDLS C++LES VVDGF KL+T+ V C LKSIPPLK D+LET+DLSCC+ L+SF
Sbjct: 74 LDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCCYSLQSFS 253
Query: 530 LVVDGILGKIKTLNVESCHNLRSIPPLKLDSLEKLDISYCGSLES 574
LV D L K+ + + H ++ IPP L L C S+ S
Sbjct: 254 LVADR-LWKLVLDDCKELHEIKVIPP----CLRMLSAVNCTSMTS 373
>BE325283 weakly similar to GP|13897750|gb resistance-gene protein {Vigna
unguiculata}, partial (74%)
Length = 576
Score = 260 bits (665), Expect = 1e-69
Identities = 142/191 (74%), Positives = 154/191 (80%), Gaps = 3/191 (1%)
Frame = +3
Query: 68 GIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEIT 127
GIGGIGKTTLALEVYNSIV QF+CSCF EKVR+ + +GLIYLQKILLS I GE N EIT
Sbjct: 3 GIGGIGKTTLALEVYNSIVHQFQCSCFFEKVRDFKE-SGLIYLQKILLSQIVGETNMEIT 179
Query: 128 SVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISS 187
SV +G+SIL++RL +KKVLLLLDDVDK EQLKAIAGSS WF GSRVIITTR K LL
Sbjct: 180 SVRQGVSILQQRLHQKKVLLLLDDVDKDEQLKAIAGSSEWFGLGSRVIITTRDKRLLTYH 359
Query: 188 GVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKND 247
G+ER YEVKGLND DAFDLVGWK LKN SP YKDVLLEQK GRE +AN+L RLK LK D
Sbjct: 360 GIERRYEVKGLNDADAFDLVGWKALKNYYSPSYKDVLLEQKQGRELNANELCRLKYLKKD 539
Query: 248 ---EGYANVLK 255
YANV K
Sbjct: 540 VRFSSYANVYK 572
>BG646177 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (17%)
Length = 708
Score = 221 bits (563), Expect = 1e-57
Identities = 124/262 (47%), Positives = 163/262 (61%)
Frame = +3
Query: 98 VRENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQ 157
VRE S K+GL LQ+ LLS G + + V EGI I+K+RL KKVLL+LDDVD+ +Q
Sbjct: 3 VREISAKHGLEDLQEKLLSKTVG-LSVKFGHVSEGIPIIKERLRLKKVLLILDDVDELKQ 179
Query: 158 LKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCS 217
LK +AG NW GSRV++TTR K+LL G+ER YE+ GLN E+A +L+ WK KN+
Sbjct: 180 LKVLAGDPNWLGHGSRVVVTTRDKHLLACHGIERTYELDGLNKEEALELLKWKAFKNN-- 353
Query: 218 PRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSN 277
K D Y ++L R V YASG PLALEV+GS
Sbjct: 354 ---------------------------KVDSSYEHILNRAVTYASGLPLALEVVGSSLFG 452
Query: 278 KTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILH 337
K ++ LDRY+R+PH + L++S+DSL+++E+ VFLDIACCF+G+ L VE IL+
Sbjct: 453 KHKDEWKSTLDRYERIPHKEVLKILKVSFDSLEKDEQSVFLDIACCFRGYILAEVEDILY 632
Query: 338 AHHGHTMKDQINVLVEKYLIKI 359
AH+G MK I VL+EK LIKI
Sbjct: 633 AHYGECMKYHIRVLIEKCLIKI 698
>TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (22%)
Length = 1736
Score = 217 bits (553), Expect(2) = 4e-57
Identities = 126/298 (42%), Positives = 172/298 (57%)
Frame = +1
Query: 6 LLDRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVG 65
LLD + Y +E IE IVE + I V L + VG++ + Q V LLL+ SD+ ++ VG
Sbjct: 664 LLDMNRYEYELIENIVEHISDRINRVFLHVAKYPVGLQSRVQQVKLLLDEESDEGVNMVG 843
Query: 66 ILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTE 125
+ G +GK+TLA +YN I QFE CFL VRENS + L +LQK LLS + N +
Sbjct: 844 LYGTRALGKSTLAKAIYNFIADQFEGVCFLHNVRENSARKNLKHLQKELLSKTV-QLNIK 1020
Query: 126 ITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLI 185
+ V EGI I+K+RL KK+LL+LDDVD+ +QL+A+AG +WF GSRVIITTR K+LL
Sbjct: 1021LRDVSEGIPIIKERLCRKKILLILDDVDQLDQLEALAGGLDWFGPGSRVIITTRDKHLLT 1200
Query: 186 SSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLK 245
G+ER Y V+GL ++A +L+ W KN+ P
Sbjct: 1201CHGIERTYAVRGLYGKEALELLRWTAFKNNKVP--------------------------- 1299
Query: 246 NDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQ 303
Y +VL R V+Y SG PL LE++GS+ K IE + LD Y R+P+ IQ L+
Sbjct: 1300--PSYEDVLNRAVSYGSGIPLVLEIVGSNLFGKNIEVWKNTLDGYDRIPNKEIQKILR 1467
Score = 23.5 bits (49), Expect(2) = 4e-57
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +2
Query: 312 EEKIVFLDIACCF 324
EE+ VFLDIAC F
Sbjct: 1466 EEQNVFLDIACFF 1504
>AL381999 similar to GP|18033111|gb functional candidate resistance protein
KR1 {Glycine max}, partial (4%)
Length = 451
Score = 207 bits (528), Expect = 1e-53
Identities = 104/150 (69%), Positives = 115/150 (76%)
Frame = +3
Query: 740 IPPCLRMLSAVNCTSLTSSCTSKLLNQELHKAGNTWFCLPRVPKIPEWFDHKYEAGLSIS 799
IPPCLR LSAVNC LTSSC S LLNQ+LH+AGNT FCLPR KIPEWFDH+ EAG+S+S
Sbjct: 3 IPPCLRELSAVNC-KLTSSCKSNLLNQKLHEAGNTRFCLPRA-KIPEWFDHQCEAGMSVS 176
Query: 800 FWFRNKFPAIALCVVSPLTWDGSRRHSIRVIINGNTFIYTDGLKMGTKSPLNMYHLHLFH 859
FWF NKFP+IAL VVS TW G H RVIIN NTF YT G K+ S + YHLHLFH
Sbjct: 177 FWFCNKFPSIALGVVSAYTW-GYLEHPARVIINDNTFFYTHGRKIDRCSRPDTYHLHLFH 353
Query: 860 MKMENFNDNMEKVLFENMWNHAEVDFGLPF 889
M++E FN NM+K L EN WNHAEVDFG PF
Sbjct: 354 MQVEYFNGNMDKALLENKWNHAEVDFGFPF 443
>TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative resistance
protein {Glycine max}, partial (8%)
Length = 2252
Score = 206 bits (525), Expect = 3e-53
Identities = 133/352 (37%), Positives = 196/352 (54%), Gaps = 6/352 (1%)
Frame = +1
Query: 43 EHQKQHVTLLLNVG---SDDSIHKVGILGIGGIGKTTLALEVYN-SIVSQFECSCFLEKV 98
+H+ + V LN DD + VGI G+ GIGKTTLA VY+ ++F+ CF + V
Sbjct: 598 DHRVEKVMRYLNSSPRSDDDGVCLVGICGVPGIGKTTLARGVYHFGGGTEFDSCCFFDNV 777
Query: 99 RENSDKNGLIYLQKILLSHIFGEKN-TEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQ 157
E K+GL++LQ++LLS I G N T +V E + +K L +KKV L+L+DV E
Sbjct: 778 GEYVKKHGLVHLQQMLLSAIVGHNNSTMFENVDERVWKIKHMLNQKKVFLVLEDVHDSEV 957
Query: 158 LKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCS 217
LKAI S +F GS+VIIT R K L G++RIYEV+ +N +AF L+
Sbjct: 958 LKAIVKLSTFFGSGSKVIITAREKCFLEFHGIKRIYEVERMNKTEAFQLL---------- 1107
Query: 218 PRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSN 277
L+ + + +L+ + YASGHP LE+IGS+ S
Sbjct: 1108-------------------NLKAFDSMNISPCHVTILEGLETYASGHPFILEMIGSYLSG 1230
Query: 278 KTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILH 337
K++E+C AL +YK + + I+ LQ+S+D+L++ ++ + + IA + +L MVE +LH
Sbjct: 1231KSMEECESALHQYKEISNRDIKKILQVSFDALEKSQQNMLIHIALHLREQELEMVENLLH 1410
Query: 338 AHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQEAP-EDPG 388
+G K I VL+ K LIKINE+G+V +H L +DM VR + P ED G
Sbjct: 1411RKYGVCPKYDIRVLLNKSLIKINENGHVIVHVLTQDM----VRDDIPVEDLG 1554
>BG645689 similar to GP|18033111|g functional candidate resistance protein
KR1 {Glycine max}, partial (14%)
Length = 746
Score = 176 bits (447), Expect(2) = 3e-51
Identities = 93/158 (58%), Positives = 117/158 (73%)
Frame = +3
Query: 250 YANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSL 309
Y +L RVV YASG PLALEV+GS+ K IE+ LD Y+R+P+ IQ L +S+++L
Sbjct: 18 YEGILNRVVTYASGLPLALEVVGSNLFGKDIEKWKSLLDEYERIPNKEIQKILIVSFNNL 197
Query: 310 QEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHD 369
E E+ VFLDIACCFKG+ L VE IL AH+G+ MK I LV+K LIKI S VTLHD
Sbjct: 198 GEYEQSVFLDIACCFKGYSLDEVEYILCAHYGYCMKYHIGKLVDKSLIKIQLS-RVTLHD 374
Query: 370 LVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENT 407
L+E MGKEIVR+E+ +PGKR+RLWF EDI++VL+ENT
Sbjct: 375 LIEIMGKEIVRKESVIEPGKRTRLWFCEDIVRVLKENT 488
Score = 44.7 bits (104), Expect(2) = 3e-51
Identities = 26/56 (46%), Positives = 36/56 (63%), Gaps = 3/56 (5%)
Frame = +2
Query: 408 GTSKIEMIHFD--GVIEV-QWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRV 460
GT E+IH D + EV W+G+AFKKM+ LKTL+ + FS+ P + P+ LRV
Sbjct: 569 GTGNTEIIHLDFSSIKEVVDWNGKAFKKMKILKTLVIKSG-HFSKAPVYFPSPLRV 733
>BF648630 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (13%)
Length = 654
Score = 187 bits (474), Expect = 2e-47
Identities = 104/236 (44%), Positives = 140/236 (59%), Gaps = 6/236 (2%)
Frame = +1
Query: 170 KGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKY 229
+GSRVIITTR K+LL S G+ +IYE GLN E A +L+ K K+
Sbjct: 7 RGSRVIITTRDKHLLSSHGITKIYEAYGLNKEQALELLRTKAFKSK-------------- 144
Query: 230 GRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDR 289
KND Y +L R + YASG PLALEV+GS+ + +C LD+
Sbjct: 145 ---------------KNDSSYDYILNRAIKYASGLPLALEVVGSNLFGMSTTECESTLDK 279
Query: 290 YKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQIN 349
Y+R+P IQ L++S+D+L EE++ VFLDIAC F + VE IL H+GH +K +
Sbjct: 280 YERIPPEDIQKILKVSFDALDEEQQSVFLDIACFFNWCESAYVEEILEYHYGHCIKSHLR 459
Query: 350 VLVEKYLIKINESGN------VTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDI 399
LV+K LIK + + VTLHDL+EDMGKEIVR E+ ++PG+RSRLW+ +DI
Sbjct: 460 ALVDKSLIKTSIQRHGMKFELVTLHDLLEDMGKEIVRHESIKEPGERSRLWYHDDI 627
>BE319124 homologue to GP|18033111|gb functional candidate resistance protein
KR1 {Glycine max}, partial (1%)
Length = 522
Score = 178 bits (452), Expect = 7e-45
Identities = 86/130 (66%), Positives = 99/130 (76%)
Frame = +2
Query: 785 PEWFDHKYEAGLSISFWFRNKFPAIALCVVSPLTWDGSRRHSIRVIINGNTFIYTDGLKM 844
PEWFDH A LS SFWFRNKFPAIALCVV T S+R +RV+INGNTF YT K+
Sbjct: 8 PEWFDHHCLARLSFSFWFRNKFPAIALCVVCSSTLHDSQR-PVRVVINGNTFFYTHDSKI 184
Query: 845 GTKSPLNMYHLHLFHMKMENFNDNMEKVLFENMWNHAEVDFGLPFQKSGIHVSKEKSNMK 904
S +MYHLHLFHM+MENFN+NM+K L EN WNHAE+DFGL F +SGIHV KE+S+M
Sbjct: 185 DRSSRPDMYHLHLFHMQMENFNENMDKALSENKWNHAELDFGLSFLESGIHVLKERSSMN 364
Query: 905 DIRFTNPEVD 914
DIRF NP+ D
Sbjct: 365 DIRFINPKYD 394
>TC78792 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1344
Score = 177 bits (448), Expect = 2e-44
Identities = 100/201 (49%), Positives = 130/201 (63%)
Frame = +1
Query: 7 LDRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGI 66
L R+GY EFIEKIV+ + I V L D+ VGVE + V L++VGS+ +GI
Sbjct: 703 LGRNGYECEFIEKIVKYISNKINHVPLHVADYPVGVESRVLKVNSLMDVGSNGEAQMIGI 882
Query: 67 LGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEI 126
G G+GKTTLA VYN I QF+ CFL VRENS K GL +L +LS + E ++
Sbjct: 883 YGNRGMGKTTLARAVYNFIADQFDGLCFLHDVRENSAKYGLEHL*VKILSKLV-ELEVKL 1059
Query: 127 TSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLIS 186
V EGI +LK+RL KKVLL+LDDV K +QL+ +AG +WF GS+VIITTR K LL S
Sbjct: 1060GDVNEGIPVLKQRLHRKKVLLILDDVHKLKQLRVLAGGLDWFGPGSKVIITTRNKQLLAS 1239
Query: 187 SGVERIYEVKGLNDEDAFDLV 207
G+ER YE+ LN+ +A +L+
Sbjct: 1240HGIERAYEIDKLNENEALELM 1302
>TC77729 weakly similar to GP|22037342|gb|AAM90012.1 disease resistance-like
protein GS3-5 {Glycine max}, partial (72%)
Length = 973
Score = 177 bits (448), Expect = 2e-44
Identities = 88/159 (55%), Positives = 113/159 (70%), Gaps = 1/159 (0%)
Frame = +1
Query: 250 YANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSL 309
Y +L RVV YASG PLA+ IG + + +E LD Y+ +P+ IQ LQ+SYD+L
Sbjct: 13 YEEILNRVVTYASGLPLAIVTIGDNLFGRKVEDWKRILDEYENIPNKDIQRILQVSYDAL 192
Query: 310 QEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINE-SGNVTLH 368
+ +EK VFLDIACCFKG K T V+ ILHAH+GH ++ + VL EK LI E +TLH
Sbjct: 193 EPKEKSVFLDIACCFKGCKWTKVKKILHAHYGHCIEHHVGVLAEKSLIGHWEYDTQMTLH 372
Query: 369 DLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENT 407
DL+EDMGKEIVRQE+P++PG+RSRLWF +DI VL +NT
Sbjct: 373 DLIEDMGKEIVRQESPKNPGERSRLWFHDDIFDVLRDNT 489
Score = 53.1 bits (126), Expect = 5e-07
Identities = 31/58 (53%), Positives = 37/58 (63%), Gaps = 3/58 (5%)
Frame = +2
Query: 408 GTSKIEMIHFDGVI---EVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLE 462
GT IEMI+ + E +WDG AF KM NLKTLI +D FS P +LP+SLR LE
Sbjct: 653 GTENIEMIYLKYGLTARETEWDGMAFNKMTNLKTLII-DDYKFSGGPGYLPSSLRYLE 823
>BF632546 similar to GP|18033111|gb functional candidate resistance protein
KR1 {Glycine max}, partial (8%)
Length = 672
Score = 174 bits (442), Expect = 1e-43
Identities = 92/135 (68%), Positives = 110/135 (81%), Gaps = 5/135 (3%)
Frame = +2
Query: 336 LHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWF 395
LHAH+G+ MKD I+VLVEK LIKI+ SGNVTLHDL+EDMGKEIVR+E+PEDPGKR+RLW
Sbjct: 77 LHAHYGNIMKDHIDVLVEKSLIKISVSGNVTLHDLIEDMGKEIVRRESPEDPGKRTRLWA 256
Query: 396 SEDIIQVLEENTGTSKIEMIH--FDGVIEVQ---WDGEAFKKMENLKTLIFSNDVFFSEN 450
EDI +V +ENTGTS I++IH FD IE + DG+AFKKM+NL+TLIFS V FSE
Sbjct: 257 YEDIKKVFKENTGTSTIKIIHFQFDPWIEKKKDASDGKAFKKMKNLRTLIFSTPVCFSET 436
Query: 451 PKHLPNSLRVLECRN 465
+H+PNSLRVLE N
Sbjct: 437 SEHIPNSLRVLEYSN 481
>TC77732 similar to GP|9965107|gb|AAG09953.1| resistance protein MG63
{Glycine max}, partial (20%)
Length = 765
Score = 173 bits (439), Expect = 2e-43
Identities = 109/266 (40%), Positives = 153/266 (56%), Gaps = 9/266 (3%)
Frame = +2
Query: 287 LDRYKRVPHNMIQTTLQLSYDSLQEE-EKIVFLDIACCFKGWKLTMVEGILHAHHGHTMK 345
LD Y+ +P IQ LQ+SYD+ + + + + KG K T V+ ILHAH+GH ++
Sbjct: 8 LDEYENIPDKDIQRILQVSYDAFEGKGSECLSRHCLLLSKGCKWTKVKKILHAHYGHCIE 187
Query: 346 DQINVLVEKYLIKINESGN-VTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLE 404
+ VL EK LI E VTLHDL+EDMGKE+VRQE+P+ PG+RSRLWF +DI+ VL
Sbjct: 188 HHVGVLAEKCLIGHWEYDTYVTLHDLIEDMGKEVVRQESPKKPGERSRLWFRDDIVNVLR 367
Query: 405 ENTGTSKIEMIHFDGVI---EVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRV- 460
+N GT IEMI+F E +WDG A +KM NLKTLI + +FS P +LP+SLR
Sbjct: 368 DNMGTGNIEMIYFKFAFTARETEWDGMACEKMTNLKTLIIEDGNYFSGGPGYLPSSLRYW 547
Query: 461 --LECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLK-LDSLETM 517
+ C L CI+ + F++ ++ +++ C L IP + L +LE
Sbjct: 548 KWISC-----PLKSLSCISSKEFNY--------MKVLTLDFCEYLTHIPDVSGLPNLEKC 688
Query: 518 DLSCCFRLESFPLVVDGILGKIKTLN 543
CF L + P + G L K++ LN
Sbjct: 689 SFRYCFSLITIPSSI-GHLNKLEILN 763
>AW684198 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (12%)
Length = 670
Score = 166 bits (419), Expect = 5e-41
Identities = 92/177 (51%), Positives = 120/177 (66%), Gaps = 9/177 (5%)
Frame = +3
Query: 316 VFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESG------NVTLHD 369
VFLDIACCFKG E IL AH+GH +K + VL EK L+KI+ + +VTLHD
Sbjct: 3 VFLDIACCFKGCGWKEFEDILRAHYGHCIKHHLGVLAEKSLVKISSTSYSGSINHVTLHD 182
Query: 370 LVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE---VQWD 426
+EDMGKE+VRQE+P++PG+RSRLW +DI+ VL+ENTGT KIEMI+ + E +
Sbjct: 183 FIEDMGKEVVRQESPKEPGERSRLWCQDDIVNVLKENTGTRKIEMIYMNFPSEEFVIDKK 362
Query: 427 GEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFSH 483
G+AFKKM LKTLI N V FS+ K+LP+SLRVL+ R + E LS ++ + H
Sbjct: 363 GKAFKKMTRLKTLIIEN-VHFSKGLKYLPSSLRVLKLRGCLSESLLSCSLSKKFQXH 530
>BF650880 weakly similar to GP|9858476|gb| resistance protein LM12 {Glycine
max}, partial (41%)
Length = 659
Score = 165 bits (418), Expect = 6e-41
Identities = 93/196 (47%), Positives = 124/196 (62%)
Frame = +3
Query: 12 YVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGG 71
Y ++ I +IV+ + I L ++ VG+ + Q V LL+ G DD +H VGI GIGG
Sbjct: 27 YEYKLIGEIVKYISNKINRQPLHVANYPVGLHSRVQEVKSLLDEGPDDGVHMVGIYGIGG 206
Query: 72 IGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGE 131
+GK+ LA +YN + QFE CFL VRENS +N L +LQ+ LL G K ++ V E
Sbjct: 207 LGKSALARAIYNFVADQFEGLCFLHDVRENSAQNNLKHLQEKLLLKTTGLK-IKLDHVCE 383
Query: 132 GISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVER 191
GI I+K+RL K+LL+LDDVD EQL A+AG +WF GSRVIITTR K+LL S +ER
Sbjct: 384 GIPIIKERLCRNKILLILDDVDDMEQLHALAGGPDWFGHGSRVIITTRDKHLLTSHDIER 563
Query: 192 IYEVKGLNDEDAFDLV 207
Y V+GL A +L+
Sbjct: 564 TYAVEGLYGTKALELL 611
>TC93052 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (12%)
Length = 787
Score = 159 bits (402), Expect = 5e-39
Identities = 84/167 (50%), Positives = 121/167 (72%), Gaps = 2/167 (1%)
Frame = +3
Query: 298 IQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLI 357
I L++SYD L+E+EK +FLDIAC F ++++ V+ +L+ H H +D I VL++K L+
Sbjct: 21 IHEILKVSYDDLEEDEKGIFLDIACFFNSYEISYVKELLYLHGFHA-EDGIQVLIDKSLM 197
Query: 358 KINESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMI-- 415
KI+ +G V +HDL++ MG+EIVRQE+ +PG+RSRLWFS+DI+QVLEEN GT +E+I
Sbjct: 198 KIDINGCVRMHDLIQSMGREIVRQESTLEPGRRSRLWFSDDIVQVLEENKGTDTVEVIIA 377
Query: 416 HFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLE 462
+ +V+W G+AF M+NLK LI N FS P+ LPNSL+VL+
Sbjct: 378 NLRKGRKVKWCGKAFGPMKNLKILIVRN-AQFSNGPQILPNSLKVLD 515
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,816,017
Number of Sequences: 36976
Number of extensions: 486391
Number of successful extensions: 3005
Number of sequences better than 10.0: 244
Number of HSP's better than 10.0 without gapping: 2598
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2823
length of query: 918
length of database: 9,014,727
effective HSP length: 105
effective length of query: 813
effective length of database: 5,132,247
effective search space: 4172516811
effective search space used: 4172516811
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC145512.9


