

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145331.13 + phase: 0 /partial
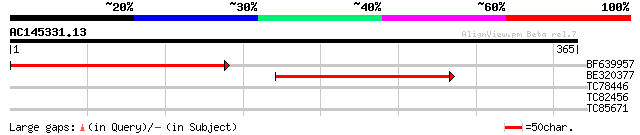
(365 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF639957 similar to GP|22128709|gb putative N-acetyl-gamma-gluta... 287 4e-78
BE320377 similar to GP|16604368|gb At2g19940/F6F22.3 {Arabidopsi... 224 5e-59
TC78446 homologue to SP|Q43814|OTC_PEA Ornithine carbamoyltransf... 32 0.30
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 32 0.51
TC85671 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 ... 28 7.3
>BF639957 similar to GP|22128709|gb putative
N-acetyl-gamma-glutamyl-phosphate reductase {Oryza
sativa (japonica cultivar-group)}, partial (27%)
Length = 683
Score = 287 bits (735), Expect = 4e-78
Identities = 141/141 (100%), Positives = 141/141 (100%)
Frame = +3
Query: 1 DLIKPQNVKLSPIKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKA 60
DLIKPQNVKLSPIKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKA
Sbjct: 237 DLIKPQNVKLSPIKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKA 416
Query: 61 GQAISSVFPHLGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSA 120
GQAISSVFPHLGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSA
Sbjct: 417 GQAISSVFPHLGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSA 596
Query: 121 DFRLRDVSEYEEWYGQPHRAP 141
DFRLRDVSEYEEWYGQPHRAP
Sbjct: 597 DFRLRDVSEYEEWYGQPHRAP 659
>BE320377 similar to GP|16604368|gb At2g19940/F6F22.3 {Arabidopsis thaliana},
partial (27%)
Length = 345
Score = 224 bits (570), Expect = 5e-59
Identities = 114/115 (99%), Positives = 115/115 (99%)
Frame = +1
Query: 172 PTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRH 231
PTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRH
Sbjct: 1 PTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRH 180
Query: 232 APEIEQGLADAAGSKVTISFTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLS 286
APEIEQGLADAAGSKVTISFTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQL+LS
Sbjct: 181 APEIEQGLADAAGSKVTISFTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQLRLS 345
>TC78446 homologue to SP|Q43814|OTC_PEA Ornithine carbamoyltransferase
chloroplast precursor (EC 2.1.3.3) (OTCase), partial
(90%)
Length = 1343
Score = 32.3 bits (72), Expect = 0.30
Identities = 17/39 (43%), Positives = 24/39 (60%)
Frame = +2
Query: 234 EIEQGLADAAGSKVTISFTPHLIPMSRGMQSTIYVEMAP 272
+++Q L DAAGSK +F H +P RG++ T V AP
Sbjct: 953 QVDQSLMDAAGSK---AFFMHCLPAERGVEVTEQVVEAP 1060
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 31.6 bits (70), Expect = 0.51
Identities = 17/47 (36%), Positives = 29/47 (61%)
Frame = +2
Query: 295 LLENGVIPRTHSVKGSNYCLFNVFPDRIPGRAIIISVIDNLVKGASG 341
+L + ++PR HSV G L + DR+P + + +++I NLVK +G
Sbjct: 383 ILRSNILPRKHSV-GLG*MLV*L*MDRVPFQELFLNIIMNLVKPKAG 520
>TC85671 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 (L1).
[Apricot] {Prunus armeniaca}, complete
Length = 1508
Score = 27.7 bits (60), Expect = 7.3
Identities = 14/36 (38%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Frame = +1
Query: 171 YPTSIQLP-LVPLIKANLIQTTNIIVDAKSGVSGAG 205
YP + LP +VP + L+ T ++VDA S + G+G
Sbjct: 286 YPVFLVLPEVVPTVPDKLLSVTCVVVDACSLLPGSG 393
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,907,304
Number of Sequences: 36976
Number of extensions: 117581
Number of successful extensions: 511
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 505
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 511
length of query: 365
length of database: 9,014,727
effective HSP length: 97
effective length of query: 268
effective length of database: 5,428,055
effective search space: 1454718740
effective search space used: 1454718740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC145331.13


