

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145330.4 + phase: 0
(257 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
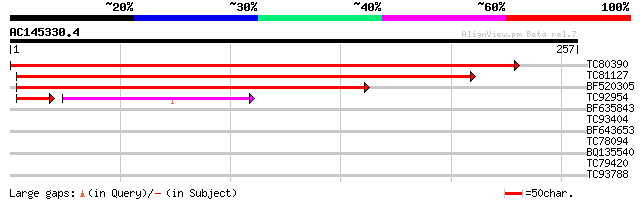
TC80390 similar to GP|22324434|dbj|BAC10351. contains ESTs AU174... 478 e-135
TC81127 similar to GP|21555141|gb|AAM63786.1 unknown {Arabidopsi... 243 5e-65
BF520305 similar to GP|21740709|em OSJNBa0086B14.2 {Oryza sativa... 202 1e-52
TC92954 similar to GP|21555141|gb|AAM63786.1 unknown {Arabidopsi... 100 1e-25
BF635843 32 0.18
TC93404 28 2.7
BF643653 similar to GP|20260210|gb unknown protein {Arabidopsis ... 28 2.7
TC78094 similar to GP|3242705|gb|AAC23757.1| unknown protein {Ar... 28 2.7
BQ135540 27 5.9
TC79420 weakly similar to GP|1354849|gb|AAB02006.1| epoxide hydr... 27 5.9
TC93788 GP|23615381|emb|CAD52372. hypothetical protein {Plasmodi... 27 7.7
>TC80390 similar to GP|22324434|dbj|BAC10351. contains ESTs AU174016(S13261)
D47643(S13261)~similar to Oryza sativa chromosome 4
OSJNBa0086B14.2~, partial (79%)
Length = 736
Score = 478 bits (1230), Expect = e-135
Identities = 231/231 (100%), Positives = 231/231 (100%)
Frame = +1
Query: 1 MEGENGRKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIF 60
MEGENGRKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIF
Sbjct: 43 MEGENGRKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIF 222
Query: 61 PPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQG 120
PPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQG
Sbjct: 223 PPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQG 402
Query: 121 KLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFD 180
KLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFD
Sbjct: 403 KLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFD 582
Query: 181 KPLIIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSICEHESKVEVD 231
KPLIIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSICEHESKVEVD
Sbjct: 583 KPLIIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSICEHESKVEVD 735
>TC81127 similar to GP|21555141|gb|AAM63786.1 unknown {Arabidopsis
thaliana}, partial (83%)
Length = 856
Score = 243 bits (620), Expect = 5e-65
Identities = 112/208 (53%), Positives = 146/208 (69%)
Frame = +3
Query: 4 ENGRKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPP 63
E GRK +ILCLHGFRTSG +KKQI KW ++ + L F D FP GKSD+EGIF PP
Sbjct: 39 EKGRKPRILCLHGFRTSGEIMKKQIHKWPQNVLDKLDLVFVDAPFPCNGKSDVEGIFDPP 218
Query: 64 YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLL 123
Y+EWFQF+K+FT YTN DEC+ Y+ +Y+I +GPFDG LGFSQGA LS L G Q +G L
Sbjct: 219 YYEWFQFNKEFTEYTNFDECLQYIEDYMIKHGPFDGLLGFSQGAILSGGLPGLQEKGVAL 398
Query: 124 KEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPL 183
+ P +KFL+ I G+KFR PS+ + AY I S+HF+GE D+LK +EL + +P+
Sbjct: 399 TKVPKVKFLIIIGGAKFRAPSVVEKAYSSQIGCPSLHFLGEHDFLKEYGKELIDSCVEPV 578
Query: 184 IIRHPQGHTVPRLDEVSTGQLQNFVAEI 211
+I HP+GHTVPRLD+ S + +F+ I
Sbjct: 579 VIHHPKGHTVPRLDDKSLNTMMSFIERI 662
>BF520305 similar to GP|21740709|em OSJNBa0086B14.2 {Oryza sativa}, partial
(62%)
Length = 655
Score = 202 bits (514), Expect = 1e-52
Identities = 92/160 (57%), Positives = 115/160 (71%)
Frame = +2
Query: 4 ENGRKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPP 63
E GRKL+ILCLHGFRTSG +KKQI KW ++ + L F D FP GKSD+EGIF PP
Sbjct: 38 EKGRKLRILCLHGFRTSGEIMKKQIHKWPQNVLDKLDLVFVDAPFPCNGKSDVEGIFDPP 217
Query: 64 YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLL 123
Y+EWFQF+K+FT YTN DEC+ Y+ +Y+I +GPFDG LGFSQGA LS L G Q +G L
Sbjct: 218 YYEWFQFNKEFTEYTNFDECLQYIEDYMIKHGPFDGLLGFSQGAILSGGLPGLQEKGVAL 397
Query: 124 KEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIG 163
+ P +KFL+ I G+KFR PS+ + AY I S+HF+G
Sbjct: 398 TKVPKVKFLIIIGGAKFRAPSVVEKAYSSQIGCPSLHFLG 517
>TC92954 similar to GP|21555141|gb|AAM63786.1 unknown {Arabidopsis
thaliana}, partial (46%)
Length = 810
Score = 99.8 bits (247), Expect(2) = 1e-25
Identities = 50/128 (39%), Positives = 65/128 (50%), Gaps = 41/128 (32%)
Frame = +1
Query: 25 KKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFEWFQFDK------------ 72
KKQI KW ++ + L F D FP GKSD+EGIF PPY+EWFQF+K
Sbjct: 103 KKQIHKWPQNVLDKLDLVFVDAPFPCNGKSDVEGIFDPPYYEWFQFNKVIIIIFYFFILS 282
Query: 73 -----------------------------DFTVYTNLDECISYLTEYIIANGPFDGFLGF 103
+FT YTN DEC+ Y+ +Y+I +GPFDG LGF
Sbjct: 283 *FGYIRRFYLLFVCLC*HVFVVQQQQ*QQEFTEYTNFDECLQYIEDYMIKHGPFDGLLGF 462
Query: 104 SQGATLSA 111
SQ +L++
Sbjct: 463 SQVRSLAS 486
Score = 33.5 bits (75), Expect(2) = 1e-25
Identities = 14/17 (82%), Positives = 15/17 (87%)
Frame = +2
Query: 4 ENGRKLKILCLHGFRTS 20
E GRK +ILCLHGFRTS
Sbjct: 38 EKGRKPRILCLHGFRTS 88
>BF635843
Length = 458
Score = 32.3 bits (72), Expect = 0.18
Identities = 14/50 (28%), Positives = 21/50 (42%)
Frame = +2
Query: 8 KLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIE 57
K ILCLHG+ + + W + S+ F D F G D++
Sbjct: 209 KKVILCLHGYLQNSEVFSSKAGSWRKGLKSRAEFHFVDAPFLVDGDQDVK 358
>TC93404
Length = 948
Score = 28.5 bits (62), Expect = 2.7
Identities = 18/71 (25%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Frame = +2
Query: 181 KPLIIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSICEHESKVEVDGTN--GDKG 238
K L++RH + +++ + L++ + E + Q K S C+H K++V + D+
Sbjct: 386 KELVLRHVNRLNLILIEKGALISLEHIILEKIPQLKEIPSGCKHLDKLKVIDLSDMSDEF 565
Query: 239 VNGVEINQGNA 249
VN +E ++G++
Sbjct: 566 VNSIEPDKGHS 598
>BF643653 similar to GP|20260210|gb unknown protein {Arabidopsis thaliana},
partial (22%)
Length = 574
Score = 28.5 bits (62), Expect = 2.7
Identities = 10/35 (28%), Positives = 20/35 (56%)
Frame = +3
Query: 64 YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFD 98
YF F ++TV++ L+ECI ++ + ++ D
Sbjct: 189 YFRKNPFGGEYTVFSGLEECIRFIANFKLSEDEID 293
>TC78094 similar to GP|3242705|gb|AAC23757.1| unknown protein {Arabidopsis
thaliana}, partial (92%)
Length = 2006
Score = 28.5 bits (62), Expect = 2.7
Identities = 10/35 (28%), Positives = 20/35 (56%)
Frame = +1
Query: 64 YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFD 98
YF F ++TV++ L+ECI ++ + ++ D
Sbjct: 196 YFRKNPFGGEYTVFSGLEECIRFIANFKLSEDEID 300
>BQ135540
Length = 726
Score = 27.3 bits (59), Expect = 5.9
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = -1
Query: 128 PIKFLVSISGSKFRDPSICDVAYKDPIKAKSVH 160
PI++ +SIS KF+D S VA P+K +++
Sbjct: 177 PIQYTLSISSVKFQDGSTDFVAVNVPVKMPTLY 79
>TC79420 weakly similar to GP|1354849|gb|AAB02006.1| epoxide hydrolase
{Nicotiana tabacum}, partial (90%)
Length = 1433
Score = 27.3 bits (59), Expect = 5.9
Identities = 9/33 (27%), Positives = 20/33 (60%)
Frame = -2
Query: 54 SDIEGIFPPPYFEWFQFDKDFTVYTNLDECISY 86
S++ + P P +F+FDK ++ +L+ ++Y
Sbjct: 313 SELNTVMPKPRLLYFRFDKSTFIFQSLETTLTY 215
>TC93788 GP|23615381|emb|CAD52372. hypothetical protein {Plasmodium
falciparum 3D7}, partial (3%)
Length = 568
Score = 26.9 bits (58), Expect = 7.7
Identities = 15/28 (53%), Positives = 16/28 (56%)
Frame = -2
Query: 10 KILCLHGFRTSGSFIKKQISKWDPSIFS 37
K L GF TS SFI IS +D IFS
Sbjct: 402 KYLSASGFATSISFIINSISSFDTFIFS 319
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.140 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,192,949
Number of Sequences: 36976
Number of extensions: 118677
Number of successful extensions: 453
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 453
length of query: 257
length of database: 9,014,727
effective HSP length: 94
effective length of query: 163
effective length of database: 5,538,983
effective search space: 902854229
effective search space used: 902854229
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC145330.4


