

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.12 - phase: 0
(319 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
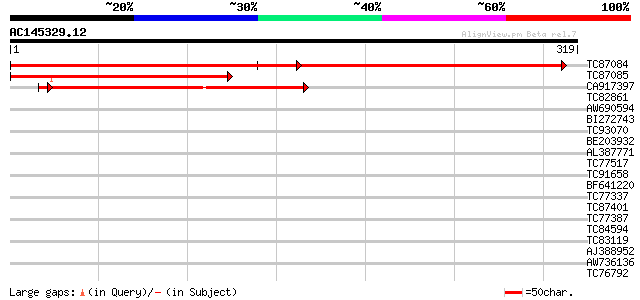
Score E
Sequences producing significant alignments: (bits) Value
TC87084 similar to GP|2529683|gb|AAC62866.1| unknown protein {Ar... 313 4e-86
TC87085 similar to PIR|T01511|T01511 hypothetical protein T10M13... 217 4e-57
CA917397 similar to GP|2529683|gb| unknown protein {Arabidopsis ... 206 8e-54
TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidop... 36 0.017
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 35 0.050
BI272743 homologue to GP|9279730|dbj formin-like protein {Arabid... 34 0.085
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 33 0.15
BE203932 similar to GP|6630459|gb|A F23N19.12 {Arabidopsis thali... 29 0.22
AL387771 weakly similar to GP|3393011|emb| Clumping factor B {St... 32 0.25
TC77517 similar to PIR|T51159|T51159 HMG protein [imported] - Ar... 32 0.25
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 32 0.25
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 32 0.32
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 32 0.42
TC87401 similar to PIR|B84587|B84587 probable glutaredoxin [impo... 31 0.55
TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19 ... 31 0.72
TC84594 similar to GP|23497092|gb|AAN36641.1 hypothetical protei... 31 0.72
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 31 0.72
AJ388952 similar to PIR|T04526|T045 hypothetical protein F16A16.... 31 0.72
AW736136 weakly similar to GP|6625522|emb| putative serine-threo... 30 0.94
TC76792 similar to GP|3264767|gb|AAC24587.1| AP2 domain containi... 30 0.94
>TC87084 similar to GP|2529683|gb|AAC62866.1| unknown protein {Arabidopsis
thaliana}, partial (81%)
Length = 2109
Score = 313 bits (803), Expect = 4e-86
Identities = 163/165 (98%), Positives = 164/165 (98%), Gaps = 1/165 (0%)
Frame = +3
Query: 1 MAVTTHSLTATEKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRL 60
MAVTTHSLTATEKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRL
Sbjct: 147 MAVTTHSLTATEKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRL 326
Query: 61 DQALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSG-SSSSSSSSDSSSSREGVK 119
DQALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSG SSSSSSSSDSSSS+EGVK
Sbjct: 327 DQALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSSDSSSSKEGVK 506
Query: 120 LLSSDSPVRDQSFKCFSVSDLKKKVMAGLCKSCEKEGQWRYLVLG 164
LLSSDSPVRDQSFKCFSVSDLKKKVMAGLCKSCEKEGQWRYLVLG
Sbjct: 507 LLSSDSPVRDQSFKCFSVSDLKKKVMAGLCKSCEKEGQWRYLVLG 641
Score = 305 bits (781), Expect = 2e-83
Identities = 154/174 (88%), Positives = 158/174 (90%)
Frame = +1
Query: 140 LKKKVMAGLCKSCEKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWS 199
L++K+ K K+G GEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWS
Sbjct: 568 LRRKLWQVCVKVVRKKGNGDTWFWGEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWS 747
Query: 200 DDSFPLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRAAALAALDAGLY 259
DDSFPLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRAAALAALDAGLY
Sbjct: 748 DDSFPLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRAAALAALDAGLY 927
Query: 260 SEAIRHFSKIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCNRTLSLDP 313
SEAIRHFSKIVDGRRAAPQGFLAECYMHRASAHRS GRIAESIADCNRTLSLDP
Sbjct: 928 SEAIRHFSKIVDGRRAAPQGFLAECYMHRASAHRSAGRIAESIADCNRTLSLDP 1089
>TC87085 similar to PIR|T01511|T01511 hypothetical protein T10M13.11 -
Arabidopsis thaliana, partial (12%)
Length = 639
Score = 217 bits (553), Expect = 4e-57
Identities = 125/168 (74%), Positives = 125/168 (74%), Gaps = 43/168 (25%)
Frame = +3
Query: 1 MAVTTHSLTATEKKHWWLTNRK-------------------------------------- 22
MAVTTHSLTATEKKHWWLTNRK
Sbjct: 135 MAVTTHSLTATEKKHWWLTNRKVTFSHNMIFSLNHPS**CFLCVLLNVVCISSSLLRSNR 314
Query: 23 -----IVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSLLYLRRF 77
IVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSLLYLRRF
Sbjct: 315 KTAFSIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSLLYLRRF 494
Query: 78 KDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
KDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS
Sbjct: 495 KDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 638
>CA917397 similar to GP|2529683|gb| unknown protein {Arabidopsis thaliana},
partial (19%)
Length = 484
Score = 206 bits (525), Expect(2) = 8e-54
Identities = 111/149 (74%), Positives = 128/149 (85%), Gaps = 2/149 (1%)
Frame = +3
Query: 22 KIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSLLYLRRFKDVA 81
++ EKYIKDAR+LIATQE++EI+SA++LLD+ALAISP DQALE++ARSLLYLRRFKDVA
Sbjct: 39 RLCEKYIKDARTLIATQERNEIVSAISLLDSALAISPSSDQALEMKARSLLYLRRFKDVA 218
Query: 82 DMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLL-SSDSPVRDQS-FKCFSVSD 139
+MLQDYIPSL+M N+D S SSSS SS SREGVKLL SSDS DQS FKCFSVSD
Sbjct: 219 NMLQDYIPSLKMANDDSGSVSSSSDGSS-QQLSREGVKLLSSSDSSAPDQSYFKCFSVSD 395
Query: 140 LKKKVMAGLCKSCEKEGQWRYLVLGEACC 168
LKK+VMAGLCKS + EG WRYLVLG+ACC
Sbjct: 396 LKKRVMAGLCKSYDNEGYWRYLVLGKACC 482
Score = 21.6 bits (44), Expect(2) = 8e-54
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = +2
Query: 17 WLTNRKIV 24
WLTNRKIV
Sbjct: 23 WLTNRKIV 46
>TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidopsis
thaliana}, partial (29%)
Length = 750
Score = 36.2 bits (82), Expect = 0.017
Identities = 21/67 (31%), Positives = 37/67 (54%)
Frame = +2
Query: 247 RAAALAALDAGLYSEAIRHFSKIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCN 306
+A AA +G +S AIRHFS+ +D +P + Y +R++A+ S +++ D
Sbjct: 98 KAKGNAAFSSGDFSTAIRHFSEAID---LSPTNHV--LYSNRSAAYASLQNYTDALTDAK 262
Query: 307 RTLSLDP 313
+T+ L P
Sbjct: 263 KTVELKP 283
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 34.7 bits (78), Expect = 0.050
Identities = 23/51 (45%), Positives = 26/51 (50%)
Frame = -3
Query: 88 IPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVS 138
I S+ ++ SS SSSSSSSS SSSS SS S S C S S
Sbjct: 262 ILSISSSSPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSSCSSSS 110
Score = 31.2 bits (69), Expect = 0.55
Identities = 24/59 (40%), Positives = 27/59 (45%), Gaps = 1/59 (1%)
Frame = -3
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKC-FSVSDLKKKVMAGLCKS 151
++ SS SSSSSSSS SSSS SS S S C S S + LC S
Sbjct: 220 SSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSSCSSSSSSSCSSSSSSTSPSSSSSLCSS 44
Score = 27.3 bits (59), Expect = 8.0
Identities = 18/44 (40%), Positives = 20/44 (44%)
Frame = -3
Query: 99 SSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKK 142
SS SSSSSSSS S SS SS S S S ++
Sbjct: 163 SSSSSSSSSSSSSCSSSSSSSCSSSSSSTSPSSSSSLCSSTFRR 32
>BI272743 homologue to GP|9279730|dbj formin-like protein {Arabidopsis
thaliana}, partial (2%)
Length = 370
Score = 33.9 bits (76), Expect = 0.085
Identities = 25/63 (39%), Positives = 33/63 (51%), Gaps = 1/63 (1%)
Frame = -3
Query: 90 SLRM-TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKKKVMAGL 148
S+RM + DP S SSSS+S+S +SSS + SS + +F C S S A L
Sbjct: 236 SIRM*SISDPLSPSSSSASNSSTSSSTSFISSCSSSINIIFSTFSCTSFSS-----TAAL 72
Query: 149 CKS 151
C S
Sbjct: 71 CSS 63
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 33.1 bits (74), Expect = 0.15
Identities = 17/39 (43%), Positives = 25/39 (63%)
Frame = -3
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSF 132
++ SS SSSSSSSS SSSS + + +SS +++F
Sbjct: 317 SSSSSSSSSSSSSSSSSSSSSSKSISSISSSESYGERNF 201
Score = 31.2 bits (69), Expect = 0.55
Identities = 18/29 (62%), Positives = 19/29 (65%)
Frame = -3
Query: 97 DPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
+PSS SSSSSSSS SSSS SS S
Sbjct: 341 NPSSSSSSSSSSSSSSSSSSSSSSSSSSS 255
Score = 30.8 bits (68), Expect = 0.72
Identities = 18/32 (56%), Positives = 21/32 (65%)
Frame = -3
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
++ SS SSSSSSSS SSSS K +SS S
Sbjct: 326 SSSSSSSSSSSSSSSSSSSSSSSSSKSISSIS 231
Score = 30.0 bits (66), Expect = 1.2
Identities = 20/50 (40%), Positives = 26/50 (52%)
Frame = -3
Query: 90 SLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSD 139
SL + + ++ SSSSSSSS SSSS SS S +S S S+
Sbjct: 371 SLALRSVGSANPSSSSSSSSSSSSSSSSSSSSSSSSSSSSKSISSISSSE 222
Score = 30.0 bits (66), Expect = 1.2
Identities = 19/46 (41%), Positives = 24/46 (51%)
Frame = -3
Query: 82 DMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPV 127
++L + S+ N SS SSSSSSSS SSSS SS +
Sbjct: 380 NLLSLALRSVGSANPSSSSSSSSSSSSSSSSSSSSSSSSSSSSKSI 243
>BE203932 similar to GP|6630459|gb|A F23N19.12 {Arabidopsis thaliana},
partial (10%)
Length = 507
Score = 28.9 bits (63), Expect(2) = 0.22
Identities = 20/50 (40%), Positives = 25/50 (50%)
Frame = +3
Query: 77 FKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSP 126
F V L+ + P + SS SSSSSSSS S S + +L S SP
Sbjct: 267 FNHVVKNLKIFFP---IDTSPGSSSSSSSSSSSPSCSFSTSLTMLPSSSP 407
Score = 22.3 bits (46), Expect(2) = 0.22
Identities = 12/33 (36%), Positives = 17/33 (51%)
Frame = +2
Query: 11 TEKKHWWLTNRKIVEKYIKDARSLIATQEQSEI 43
++KK W LT R + Y+K R + Q EI
Sbjct: 161 SQKKTWKLTQRLWLLSYLKGMRMYLIM*CQREI 259
>AL387771 weakly similar to GP|3393011|emb| Clumping factor B {Staphylococcus
aureus}, partial (5%)
Length = 473
Score = 32.3 bits (72), Expect = 0.25
Identities = 17/35 (48%), Positives = 21/35 (59%)
Frame = +3
Query: 90 SLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSD 124
S R + D SS SSS S SD+SSS EG++ D
Sbjct: 168 SSRSRSSDGSSSESSSDSESDTSSSSEGLESTDED 272
>TC77517 similar to PIR|T51159|T51159 HMG protein [imported] - Arabidopsis
thaliana, partial (70%)
Length = 907
Score = 32.3 bits (72), Expect = 0.25
Identities = 22/53 (41%), Positives = 29/53 (54%)
Frame = -1
Query: 71 LLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSS 123
LLY + K++ ++ +I S + SS SSSSSSSS SSS L SS
Sbjct: 709 LLY*KIHKNLPPPMKHHISSYAHKSHSSSSSSSSSSSSSSSSSWWSSSPLASS 551
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 32.3 bits (72), Expect = 0.25
Identities = 24/53 (45%), Positives = 26/53 (48%)
Frame = -2
Query: 88 IPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDL 140
+P L + PSS SSSSSSSS SSSS SS SP S S L
Sbjct: 213 LPLLLLLFPFPSSSSSSSSSSSSSSSSSSSSS--SSSSPSSSSSSTSISSKSL 61
Score = 30.0 bits (66), Expect = 1.2
Identities = 18/34 (52%), Positives = 21/34 (60%)
Frame = -2
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPV 127
++ SS SSSSS SS SSS+ K LS SPV
Sbjct: 144 SSSSSSSSSSSSSPSSSSSSTSISSKSLSPASPV 43
Score = 28.9 bits (63), Expect = 2.7
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = -2
Query: 99 SSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFS 136
SS SSSSSSSS SSSS SS S S K S
Sbjct: 171 SSSSSSSSSSSSSSSSSSSSSSPSSSSSSTSISSKSLS 58
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 32.0 bits (71), Expect = 0.32
Identities = 21/45 (46%), Positives = 22/45 (48%)
Frame = -2
Query: 89 PSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFK 133
PSL T SS SSSSSSS SSSS SS S S +
Sbjct: 486 PSLPXTXXSSSSSXSSSSSSSPSSSSSSSSSSSSSSSSSSSSSIE 352
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 31.6 bits (70), Expect = 0.42
Identities = 18/33 (54%), Positives = 20/33 (60%)
Frame = -2
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSP 126
++ P S SSSSSSSS SSSS SS SP
Sbjct: 688 SSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSP 590
>TC87401 similar to PIR|B84587|B84587 probable glutaredoxin [imported] -
Arabidopsis thaliana, partial (63%)
Length = 914
Score = 31.2 bits (69), Expect = 0.55
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 4/60 (6%)
Frame = +3
Query: 89 PSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPV----RDQSFKCFSVSDLKKKV 144
P ++ PS SSSSSSSS S E +K S +PV + C V L KK+
Sbjct: 204 PRAMSSSSSPSPSSSSSSSSSFGSRLEETIKNTVSQNPVVVYSKSWCSYCSEVKSLFKKL 383
>TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19
{Arabidopsis thaliana}, partial (75%)
Length = 1793
Score = 30.8 bits (68), Expect = 0.72
Identities = 23/80 (28%), Positives = 38/80 (46%)
Frame = -2
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKKKVMAGLCKSCE 153
++ SS SSSSSSSS SSSS + + +R+ + +L++K M L
Sbjct: 385 SSSSSSSSSSSSSSSSSSSSSCSWFIVDEEEEHLRNNIGELLGFLELERKRMLDLWLMGF 206
Query: 154 KEGQWRYLVLGEACCHLGLM 173
++ W + G C L ++
Sbjct: 205 EKRWWEWEGYGRIECWLRVL 146
Score = 28.5 bits (62), Expect = 3.6
Identities = 19/42 (45%), Positives = 24/42 (56%), Gaps = 1/42 (2%)
Frame = -2
Query: 85 QDYIPSLRM-TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
Q + P + + T+ + SG SSSSSSS SSSS SS S
Sbjct: 454 QSFSPWISLHTSLNGGSGDSSSSSSSSSSSSSSSSSSSSSSS 329
>TC84594 similar to GP|23497092|gb|AAN36641.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (0%)
Length = 791
Score = 30.8 bits (68), Expect = 0.72
Identities = 24/52 (46%), Positives = 28/52 (53%), Gaps = 4/52 (7%)
Frame = -3
Query: 90 SLRMTNEDPSSGSSSSSSSSDSSSSREGVKLL----SSDSPVRDQSFKCFSV 137
SL T PSS SSSSSSSS S S GV L S P +++F+ SV
Sbjct: 495 SLSTTVSPPSSSSSSSSSSSPSESLLLGVPSLQFSTSDGFPRSEKTFRRNSV 340
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 30.8 bits (68), Expect = 0.72
Identities = 25/86 (29%), Positives = 40/86 (46%), Gaps = 6/86 (6%)
Frame = +3
Query: 90 SLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSD------SPVRDQSFKCFSVSDLKKK 143
+L+M+++ S S S S S S S R ++ + SD +P R S + FS +L
Sbjct: 183 NLKMSSDSRSRSRSRSRSRSRSRSPR--IRKIRSDRHSYRDAPYRRDSSRGFSRDNL--- 347
Query: 144 VMAGLCKSCEKEGQWRYLVLGEACCH 169
CK+C++ G + A CH
Sbjct: 348 -----CKNCKRPGHYARECPNVAVCH 410
>AJ388952 similar to PIR|T04526|T045 hypothetical protein F16A16.160 -
Arabidopsis thaliana, partial (24%)
Length = 591
Score = 30.8 bits (68), Expect = 0.72
Identities = 20/44 (45%), Positives = 24/44 (54%), Gaps = 5/44 (11%)
Frame = +3
Query: 89 PSLRMTNEDPSSGSSSSSSSSDSSSS-----REGVKLLSSDSPV 127
P ++ PS SSSSSSSS SSSS E +K S +PV
Sbjct: 198 PRAMSSSSSPSPSSSSSSSSSSSSSSFGSRLEETIKNTVSQNPV 329
>AW736136 weakly similar to GP|6625522|emb| putative serine-threonine-protein
kinase {Schizosaccharomyces pombe}, partial (3%)
Length = 642
Score = 30.4 bits (67), Expect = 0.94
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +3
Query: 89 PSLRMTNEDPSSGSSSSSSSSDSSSSREG 117
PS + N PSS SSS+SSSDS + G
Sbjct: 453 PSFSINNSQPSSTLSSSTSSSDSLRQQRG 539
>TC76792 similar to GP|3264767|gb|AAC24587.1| AP2 domain containing protein
{Prunus armeniaca}, partial (49%)
Length = 1729
Score = 30.4 bits (67), Expect = 0.94
Identities = 19/40 (47%), Positives = 24/40 (59%), Gaps = 3/40 (7%)
Frame = -1
Query: 76 RFKDVADML---QDYIPSLRMTNEDPSSGSSSSSSSSDSS 112
R KD+ D+L + + L TN SS SSSSSS S+SS
Sbjct: 445 RTKDLEDLLFLLEAKVKPLTPTNNPSSSSSSSSSSKSESS 326
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.130 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,386,181
Number of Sequences: 36976
Number of extensions: 161649
Number of successful extensions: 3852
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 1683
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2615
length of query: 319
length of database: 9,014,727
effective HSP length: 96
effective length of query: 223
effective length of database: 5,465,031
effective search space: 1218701913
effective search space used: 1218701913
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC145329.12


