

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.8 + phase: 0
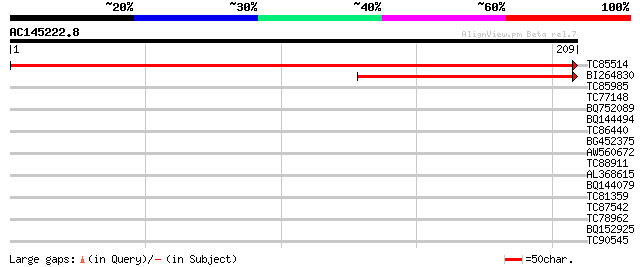
(209 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85514 homologue to SP|P32869|PSAD_CUCSA Photosystem I reaction... 426 e-120
BI264830 similar to SP|P51279|PSAD_ Photosystem I reaction cente... 114 3e-26
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 31 0.30
TC77148 ENOD20 31 0.39
BQ752089 similar to GP|14161441|gb| thiol-specific antioxidant {... 28 2.5
BQ144494 28 3.3
TC86440 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.... 27 4.3
BG452375 weakly similar to GP|20259341|gb unknown protein {Arabi... 27 5.6
AW560672 similar to GP|20259341|gb unknown protein {Arabidopsis ... 27 5.6
TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2... 27 5.6
AL368615 similar to SP|P09125|MSP8_ Merozoite surface protein CM... 27 7.4
BQ144079 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 27 7.4
TC81359 weakly similar to GP|5091614|gb|AAD39602.1| Contains a P... 26 9.6
TC87542 weakly similar to GP|22136436|gb|AAM91296.1 putative pro... 26 9.6
TC78962 similar to PIR|S38958|S38958 chorismate mutase (EC 5.4.9... 26 9.6
BQ152925 weakly similar to GP|6448504|emb| Trihydrophobin {Clavi... 26 9.6
TC90545 similar to GP|11072026|gb|AAG28905.1 F12A21.16 {Arabidop... 26 9.6
>TC85514 homologue to SP|P32869|PSAD_CUCSA Photosystem I reaction center
subunit II chloroplast precursor (Photosystem I 20 kDa
subunit), partial (77%)
Length = 870
Score = 426 bits (1094), Expect = e-120
Identities = 209/209 (100%), Positives = 209/209 (100%)
Frame = +3
Query: 1 MAMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEE 60
MAMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEE
Sbjct: 75 MAMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEE 254
Query: 61 APAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAI 120
APAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAI
Sbjct: 255 APAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAI 434
Query: 121 MREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGR 180
MREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGR
Sbjct: 435 MREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGR 614
Query: 181 QGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
QGVGQNFRSIGKNVSPIEVKFTGKQPFDV
Sbjct: 615 QGVGQNFRSIGKNVSPIEVKFTGKQPFDV 701
>BI264830 similar to SP|P51279|PSAD_ Photosystem I reaction center subunit II
(Photosystem I 16 kDa polypeptide) (PSI-D)., partial
(51%)
Length = 394
Score = 114 bits (285), Expect = 3e-26
Identities = 54/81 (66%), Positives = 63/81 (77%)
Frame = +2
Query: 129 KLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFR 188
+LARKEQCLAL +LR+K+KI YRVFPNGEVQYLHPKDGVYPEKVN GR G R
Sbjct: 5 ELARKEQCLALLTQLRTKFKIDGYIYRVFPNGEVQYLHPKDGVYPEKVNAGRTGDNTQDR 184
Query: 189 SIGKNVSPIEVKFTGKQPFDV 209
IG+NV+P +KFTGK P ++
Sbjct: 185 RIGQNVNPANIKFTGKMPAEI 247
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 31.2 bits (69), Expect = 0.30
Identities = 23/77 (29%), Positives = 32/77 (40%), Gaps = 1/77 (1%)
Frame = +3
Query: 13 PLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEA-ASVTTKEEAPAAPVGFTPP 71
P+SSP P+ + P +K+ K + +K A A E PA PVG P
Sbjct: 426 PVSSPPTASTPAVTPSAEV-PAAAPSKSKKKTKKGKKHSAPAPSPALEGPPAPPVGAPGP 602
Query: 72 ELDPNTPSPIFGGSTGG 88
LD ++P P G
Sbjct: 603 SLDASSPGPASAADESG 653
>TC77148 ENOD20
Length = 1108
Score = 30.8 bits (68), Expect = 0.39
Identities = 27/92 (29%), Positives = 34/92 (36%), Gaps = 11/92 (11%)
Frame = +3
Query: 8 SLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAP-- 65
SL +PP SP P PS PI K + S + + + S + E AP
Sbjct: 492 SLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSP 671
Query: 66 ---VGFTPPELDPN------TPSPIFGGSTGG 88
V P P+ PSP GS GG
Sbjct: 672 SDSVASLAPSSSPSDESPSPAPSPSSSGSKGG 767
>BQ752089 similar to GP|14161441|gb| thiol-specific antioxidant
{Ajellomyces capsulatus}, partial (30%)
Length = 204
Score = 28.1 bits (61), Expect = 2.5
Identities = 21/73 (28%), Positives = 29/73 (38%), Gaps = 1/73 (1%)
Frame = +1
Query: 8 SLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVG 67
S +PP S PKP PS T S++ ++S T + P+AP
Sbjct: 34 SALSPPTSKPKPPTAPS----------------TSTSSSATAGSSSSPTPRTTLPSAPPS 165
Query: 68 FTP-PELDPNTPS 79
P P P+ PS
Sbjct: 166SAPLPNSSPSFPS 204
>BQ144494
Length = 802
Score = 27.7 bits (60), Expect = 3.3
Identities = 13/49 (26%), Positives = 23/49 (46%)
Frame = -1
Query: 32 KPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSP 80
+ ++FT + +D + A + T++ AP PPE +P P P
Sbjct: 661 RTLQFTRRRA*ARTSDTRGAADADLTRQVRTVAPATKKPPEPEPTFPKP 515
>TC86440 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.1.7)
neutral - horseradish, partial (87%)
Length = 1272
Score = 27.3 bits (59), Expect = 4.3
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Frame = +2
Query: 125 PNLLKLARKEQCLALGNRLR-SKYKIKYQFYRVFPNG 160
PNL K+ RKE AL N +R ++ F+ F NG
Sbjct: 194 PNLTKIVRKEVVKALKNEMRMGASLLRLHFHDCFVNG 304
>BG452375 weakly similar to GP|20259341|gb unknown protein {Arabidopsis
thaliana}, partial (5%)
Length = 640
Score = 26.9 bits (58), Expect = 5.6
Identities = 17/54 (31%), Positives = 25/54 (45%)
Frame = +3
Query: 33 PIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGST 86
P+K T +I +TE S+ TK PAA + + + + I GGST
Sbjct: 42 PLKEVDSTKEIPIP--RTERTSMFTKRSTPAATIHIQHKKPASSVDAQIIGGST 197
>AW560672 similar to GP|20259341|gb unknown protein {Arabidopsis thaliana},
partial (4%)
Length = 556
Score = 26.9 bits (58), Expect = 5.6
Identities = 17/54 (31%), Positives = 25/54 (45%)
Frame = +3
Query: 33 PIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGST 86
P+K T +I +TE S+ TK PAA + + + + I GGST
Sbjct: 15 PLKEVDSTKEIPIP--RTERTSMFTKRSTPAATIHIQHKKPASSVDAQIIGGST 170
>TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2G14_40
{Arabidopsis thaliana}, partial (11%)
Length = 1320
Score = 26.9 bits (58), Expect = 5.6
Identities = 20/71 (28%), Positives = 29/71 (40%), Gaps = 2/71 (2%)
Frame = +3
Query: 12 PPLSSPKPWKQPSTLSFISLKPIKFTT--KTTKISAADEKTEAASVTTKEEAPAAPVGFT 69
PP SP P K P + P T K A D++T A + T K++
Sbjct: 558 PPKVSPAPAKVPKAPAPAKEAPAPAPTHKKKAPAPAPDKETPAPAPTHKKKKAPKSSPVP 737
Query: 70 PPELDPNTPSP 80
P + P +P+P
Sbjct: 738 SPAILPPSPAP 770
>AL368615 similar to SP|P09125|MSP8_ Merozoite surface protein CMZ-8
(Fragment). {Eimeria acervulina}, partial (5%)
Length = 498
Score = 26.6 bits (57), Expect = 7.4
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 6/70 (8%)
Frame = +1
Query: 18 KPWKQPSTLSFISLK-PIK-----FTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPP 71
K + Q + F L P+K F K++K +K E + V E P A P
Sbjct: 82 KKYSQKFAMDFDDLDAPVKPRVSRFAPKSSKFKP--KKEEPSLVPKSEPPPLASAKTEPQ 255
Query: 72 ELDPNTPSPI 81
E+D P+P+
Sbjct: 256 EIDLTAPTPV 285
>BQ144079 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (12%)
Length = 1071
Score = 26.6 bits (57), Expect = 7.4
Identities = 19/70 (27%), Positives = 28/70 (39%)
Frame = +3
Query: 11 TPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTP 70
TPP +P P + + + + KP+ T++ I E+P P TP
Sbjct: 627 TPP--NPPPPRSATKIRHPTAKPLPQETQSHPIP---------------ESPNTPHSHTP 755
Query: 71 PELDPNTPSP 80
P PN P P
Sbjct: 756 PPRPPNPPIP 785
>TC81359 weakly similar to GP|5091614|gb|AAD39602.1| Contains a PF|00561
alpha/beta hydrolase fold domain. {Arabidopsis
thaliana}, partial (9%)
Length = 436
Score = 26.2 bits (56), Expect = 9.6
Identities = 20/71 (28%), Positives = 29/71 (40%)
Frame = +2
Query: 11 TPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTP 70
TP SS P P S I+ P +S + S+++K +P +P P
Sbjct: 122 TPTTSSSNP--SP*FQSTITSSP------PPSLSHTSSTISSKSISSKISSPDSPA---P 268
Query: 71 PELDPNTPSPI 81
P P TP P+
Sbjct: 269 PSTSPTTPHPL 301
>TC87542 weakly similar to GP|22136436|gb|AAM91296.1 putative protein
{Arabidopsis thaliana}, partial (39%)
Length = 2234
Score = 26.2 bits (56), Expect = 9.6
Identities = 13/50 (26%), Positives = 22/50 (44%)
Frame = +2
Query: 11 TPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEE 60
+PP P P S +S ++ +P +SA + K E+ + EE
Sbjct: 401 SPPPPPPPPHIPDSLVSTVTQQPSSDVAPAVSVSATELKNESRDASNDEE 550
>TC78962 similar to PIR|S38958|S38958 chorismate mutase (EC 5.4.99.5)
precursor - Arabidopsis thaliana, partial (75%)
Length = 1318
Score = 26.2 bits (56), Expect = 9.6
Identities = 17/58 (29%), Positives = 24/58 (41%)
Frame = +2
Query: 2 AMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKE 59
A QAS P S + Q + S S KPI + + +S + SV TK+
Sbjct: 197 AKLLQASTICTPPSPSSSFHQKNGGSSFSFKPISYFPQKFNLSVKAQAASIGSVPTKK 370
>BQ152925 weakly similar to GP|6448504|emb| Trihydrophobin {Claviceps
fusiformis}, partial (13%)
Length = 614
Score = 26.2 bits (56), Expect = 9.6
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Frame = +2
Query: 37 TTKTTK-ISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTP 78
+TKTTK + + DEK T + P F PP L PN P
Sbjct: 146 STKTTKNVQSLDEK----KATNLDSFNLPPNPFFPPPLLPNNP 262
>TC90545 similar to GP|11072026|gb|AAG28905.1 F12A21.16 {Arabidopsis
thaliana}, partial (33%)
Length = 1262
Score = 26.2 bits (56), Expect = 9.6
Identities = 13/28 (46%), Positives = 16/28 (56%)
Frame = -2
Query: 141 NRLRSKYKIKYQFYRVFPNGEVQYLHPK 168
NR S K+ Y FYR+ + QYL PK
Sbjct: 1105 NRWFSS*KV*YHFYRILRSSWGQYLVPK 1022
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,190,230
Number of Sequences: 36976
Number of extensions: 85823
Number of successful extensions: 535
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 528
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 534
length of query: 209
length of database: 9,014,727
effective HSP length: 92
effective length of query: 117
effective length of database: 5,612,935
effective search space: 656713395
effective search space used: 656713395
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC145222.8


