

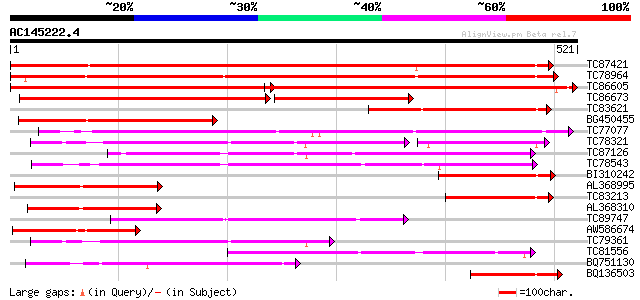
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.4 - phase: 0
(521 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87421 sugar transporter 600 e-172
TC78964 weakly similar to PIR|T00450|T00450 probable monosacchar... 531 e-151
TC86605 similar to PIR|T01853|T01853 probable hexose transport p... 453 e-128
TC86673 weakly similar to PIR|S25015|S25015 monosaccharide trans... 260 e-100
TC83621 similar to PIR|T01506|T01506 probable hexose transport p... 236 2e-62
BG450455 similar to SP|Q07423|HEX6 Hexose carrier protein HEX6. ... 212 2e-55
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 209 2e-54
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 184 7e-47
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 169 2e-42
TC78543 similar to PIR|T14545|T14545 probable sugar transporter ... 165 3e-41
BI310242 similar to GP|347853|gb|A glucose transporter {Saccharu... 163 1e-40
AL368995 similar to GP|2104547|gb| AGAA.1 {Arabidopsis thaliana}... 137 1e-32
TC83213 weakly similar to GP|16945177|emb|CAC69071. STP5 protein... 136 2e-32
AL368310 weakly similar to GP|16945177|em STP5 protein {Arabidop... 130 9e-31
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 127 1e-29
AW586674 weakly similar to SP|Q10710|STA_ Sugar carrier protein ... 121 6e-28
TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 121 7e-28
TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar tran... 117 1e-26
BQ751130 weakly similar to GP|15289796|dbj putative monosacchari... 105 4e-23
BQ136503 weakly similar to PIR|T05156|T051 probable glucose tran... 100 2e-21
>TC87421 sugar transporter
Length = 1753
Score = 600 bits (1548), Expect = e-172
Identities = 302/502 (60%), Positives = 381/502 (75%), Gaps = 3/502 (0%)
Frame = +2
Query: 1 MAGGGFATSGGG-EFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPA 59
MAGGG GG E+ +TP V I+CI+AA GGL+FGYD+G+SGGVTSM PFLKKFFPA
Sbjct: 29 MAGGGIPIGGGNKEYPGNLTPFVTITCIVAAMGGLIFGYDIGISGGVTSMDPFLKKFFPA 208
Query: 60 VYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFF 119
VYRK + + YC+YD+Q L +FTSSLYLAAL S+ AS TR GR+L+ML G
Sbjct: 209 VYRKKNKDKSTNQ-YCQYDSQTLTMFTSSLYLAALLSSLVASTITRRFGRKLSMLFGGLL 385
Query: 120 FIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVT 179
F+ G N A ++ MLIVGRILLG G+GFANQ VP++LSE+AP + RGALNI FQL++T
Sbjct: 386 FLVGALINGFANHVWMLIVGRILLGFGIGFANQPVPLYLSEMAPYKYRGALNIGFQLSIT 565
Query: 180 IGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGK 239
IGIL AN++NY KI GGWGWRLSLG A +PAL++T+G++V+ DTPNS+IERG + K
Sbjct: 566 IGILVANVLNYFFAKIKGGWGWRLSLGGAMVPALIITIGSLVLPDTPNSMIERGDRDGAK 745
Query: 240 AVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGI 299
A LK+IRG ++++ EF +L AS + +V+ +RNLL+ + Q+ MA+ + FQQFTGI
Sbjct: 746 AQLKRIRGIEDVDEEFNDLVAASEASMQVENPWRNLLQRKYRPQLTMAVLIPFFQQFTGI 925
Query: 300 NAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMF 359
N IMFYAPVLFN++GFK+DASL SAVITG VNV++T VSIY VDK GRR L LE G QM
Sbjct: 926 NVIMFYAPVLFNSIGFKDDASLMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGAQML 1105
Query: 360 LSQIVIAIILGIK--VTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPL 417
+ Q+ +A +G K + + +L + YAI VV+ +C +V+ FAWSWGPLGWL+PSE FPL
Sbjct: 1106ICQVAVAAAIGAKFGTSGNPGNLPEWYAIVVVLFICIYVAGFAWSWGPLGWLVPSEIFPL 1285
Query: 418 ETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKN 477
E RSA QSV V VNMLFTF++AQ FL MLCH KFG+FLFF+ +VL+MSI+V FL+PETK
Sbjct: 1286EIRSAAQSVNVSVNMLFTFLVAQVFLIMLCHMKFGLFLFFAFFVLVMSIYVFFLLPETKG 1465
Query: 478 IPIEEMTERVWKQHWFWKRFME 499
IPIEEM +RVWK H FW RF+E
Sbjct: 1466IPIEEM-DRVWKSHPFWSRFVE 1528
>TC78964 weakly similar to PIR|T00450|T00450 probable monosaccharide
transport protein T14N5.7 - Arabidopsis thaliana,
partial (98%)
Length = 1771
Score = 531 bits (1368), Expect = e-151
Identities = 265/509 (52%), Positives = 362/509 (71%), Gaps = 5/509 (0%)
Frame = +1
Query: 1 MAGGGFATSGGGE----FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKF 56
MAGGG G G+ +E K T +C++ A GG +FGYD+GVSGGVTSM FL+KF
Sbjct: 31 MAGGGLTNGGPGKRAHLYEHKFTAYFAFTCVVGALGGSLFGYDLGVSGGVTSMDDFLEKF 210
Query: 57 FPAVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIA 116
FP VYRK +++YCKYDNQ L LFTSSLY +AL TFFASY TR GR+ T+++
Sbjct: 211 FPDVYRKKHAHLK-ETDYCKYDNQVLTLFTSSLYFSALVMTFFASYLTRNKGRKATIIVG 387
Query: 117 GFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQL 176
F+ G NAAAQN+ LI+GR+ LG G+GF NQAVP++LSE+AP+ RGA+N LFQ
Sbjct: 388 ALSFLIGAILNAAAQNIPTLIIGRVFLGGGIGFGNQAVPLYLSEMAPASSRGAVNQLFQF 567
Query: 177 NVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLE 236
GIL ANLVNY T+KI GWR+SLGLAGIPA+L+ +G I +TPNSL+E+GRL+
Sbjct: 568 TTCAGILIANLVNYFTDKIH-PHGWRISLGLAGIPAVLMLLGGIFCAETPNSLVEQGRLD 744
Query: 237 EGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIM-AIGLEIFQQ 295
E + VL+K+RGT N++ EF +L +AS +A+ VK F+ LLK + Q+I+ A+G FQQ
Sbjct: 745 EARKVLEKVRGTKNVDAEFEDLKDASELAQAVKSPFKVLLKRKYRPQLIIGALGTPAFQQ 924
Query: 296 FTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAG 355
TG N+I+FYAPV+F + GF ++A+L+S+ IT +++T++S++ VDK G R LEAG
Sbjct: 925 LTGNNSILFYAPVIFQSSGFGSNAALFSSFITNGALLVATVISMFLVDKFGTRKFFLEAG 1104
Query: 356 VQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETF 415
+M I+ A++L ++ H +LSKG + F+VI++ FV A+ SWGPLGWL+PSE F
Sbjct: 1105FEMICCMIITAVVLAVEF-GHGKELSKGISAFLVIMIFWFVLAYGRSWGPLGWLVPSELF 1281
Query: 416 PLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPET 475
PLE RSA QS+ VCVNM+FT ++AQ FL LCH K+GIFL F G +++MS+FV FL+PET
Sbjct: 1282PLEIRSAAQSIAVCVNMIFTALVAQLFLLSLCHLKYGIFLLFGGLIVVMSVFVFFLLPET 1461
Query: 476 KNIPIEEMTERVWKQHWFWKRFMEDDNEK 504
K +PIEE+ +++ HWFWK + + ++
Sbjct: 1462KQVPIEEI-YLLFENHWFWKNIVREGTDQ 1545
>TC86605 similar to PIR|T01853|T01853 probable hexose transport protein
F9D12.17 - Arabidopsis thaliana, partial (91%)
Length = 3111
Score = 453 bits (1166), Expect = e-128
Identities = 227/290 (78%), Positives = 255/290 (87%), Gaps = 3/290 (1%)
Frame = +3
Query: 235 LEEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQ 294
L++GKAVL+KIRGTDNIEPEFLEL EASRVAKEVK FRNLLK Q++++I L IFQ
Sbjct: 1992 LDKGKAVLRKIRGTDNIEPEFLELVEASRVAKEVKHPFRNLLKRNNRPQLVISIALMIFQ 2171
Query: 295 QFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEA 354
QFTGINAIMFYAPVLFNT+GFKNDA+LYSAVITGA+NV+STIVSIY VDKLGRR LLLEA
Sbjct: 2172 QFTGINAIMFYAPVLFNTLGFKNDAALYSAVITGAINVISTIVSIYSVDKLGRRKLLLEA 2351
Query: 355 GVQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSET 414
GVQM LSQ+VIAI+LGIKV DHS++LSKGYA VV++VC FVSAFAWSWGPL WLIPSE
Sbjct: 2352 GVQMLLSQMVIAIVLGIKVKDHSEELSKGYAALVVVMVCIFVSAFAWSWGPLAWLIPSEI 2531
Query: 415 FPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPE 474
FPLETRSAGQSVTVCVN LFT VIAQAFLSMLC+FKFGIF FFSGW+L MS FV FLVPE
Sbjct: 2532 FPLETRSAGQSVTVCVNFLFTAVIAQAFLSMLCYFKFGIFFFFSGWILFMSTFVFFLVPE 2711
Query: 475 TKNIPIEEMTERVWKQHWFWKRFMEDD---NEKVSNADYPKIKNNPNSQL 521
TKN+PIEEMT+RVWKQHWFWKRF+E+D +EKV+ + P +N+ SQL
Sbjct: 2712 TKNVPIEEMTQRVWKQHWFWKRFVENDYIEDEKVTGGNSPS-RNDLVSQL 2858
Score = 392 bits (1006), Expect = e-109
Identities = 195/244 (79%), Positives = 214/244 (86%)
Frame = +2
Query: 1 MAGGGFATSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAV 60
M GGGF+ EFEAKITPI+IISCIMAATGGLMFGYDVGVSGGV SM PFLKKFFP V
Sbjct: 1289 MTGGGFSGGNDREFEAKITPIIIISCIMAATGGLMFGYDVGVSGGVASMPPFLKKFFPTV 1468
Query: 61 YRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFF 120
R+T G +SNYCKYDNQGLQLFTSSLYLA LT TFFASYTTR +GRRLTMLIAGFFF
Sbjct: 1469 LRQTTESDGSESNYCKYDNQGLQLFTSSLYLAGLTVTFFASYTTRVLGRRLTMLIAGFFF 1648
Query: 121 IAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTI 180
IAGV+ NA+AQNL MLIVGR+LLGCG+GFANQAVPVFLSEIAPSRIRGALNILFQL++T+
Sbjct: 1649 IAGVSLNASAQNLLMLIVGRVLLGCGIGFANQAVPVFLSEIAPSRIRGALNILFQLDITL 1828
Query: 181 GILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKA 240
GIL+ANLVNY TNKI G WGWR+SLGL GIPALLLT+GA +VVDTPNSLIERG L + K+
Sbjct: 1829 GILYANLVNYATNKIKGHWGWRISLGLGGIPALLLTLGAYLVVDTPNSLIERGHLGQRKS 2008
Query: 241 VLKK 244
+K
Sbjct: 2009 CSEK 2020
>TC86673 weakly similar to PIR|S25015|S25015 monosaccharide transport
protein MST1 - common tobacco, partial (65%)
Length = 1162
Score = 260 bits (664), Expect(2) = e-100
Identities = 129/230 (56%), Positives = 170/230 (73%)
Frame = +2
Query: 10 GGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAG 69
G ++ K+T VII+CIMAATGGL+FGYD GVSGGVTSM FLK+FFP+VY +
Sbjct: 59 GPSKYPGKLTFRVIITCIMAATGGLIFGYDHGVSGGVTSMDSFLKRFFPSVYEQESNLKP 238
Query: 70 LDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAA 129
+ YCK+++Q L LFTSSLY++AL + AS TR +GRR TM++ G FF++G N
Sbjct: 239 SANQYCKFNSQILTLFTSSLYISALVAGLGASSITRALGRRTTMILGGIFFVSGALLNGL 418
Query: 130 AQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVN 189
A N+AMLI GR+LLG G+G ANQAVP++LSE+AP + RGALN+ FQL++TIGI AN+ N
Sbjct: 419 AMNIAMLIAGRLLLGFGIGCANQAVPIYLSEMAPYKYRGALNMCFQLSITIGIFTANMFN 598
Query: 190 YGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGK 239
Y +KI G GWRLSLGL +PA++ +G+I + D+PNSL+ RGR EE +
Sbjct: 599 YYFSKILNGEGWRLSLGLGAVPAVVFIIGSICLPDSPNSLVTRGRHEEAR 748
Score = 124 bits (312), Expect(2) = e-100
Identities = 60/128 (46%), Positives = 88/128 (67%)
Frame = +3
Query: 244 KIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIM 303
KIRGTD+++ EF ++ AS + +VK ++ L + + Q++ AI + FQQFTG+N I
Sbjct: 762 KIRGTDDVDAEFRDIVAASEASAKVKHPWKTLQERKYRPQLVFAILIPFFQQFTGLNVIT 941
Query: 304 FYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQI 363
FYAP+LF T+GF + ASL SA I G+ +ST+VSI+ VDK GRR L LE GVQM +S I
Sbjct: 942 FYAPILFRTIGFGSQASLMSAAIIGSFKPVSTLVSIFVVDKFGRRALFLEGGVQMLISXI 1121
Query: 364 VIAIILGI 371
++ + + +
Sbjct: 1122IMTVAIAV 1145
>TC83621 similar to PIR|T01506|T01506 probable hexose transport protein
T10M13.6 - Arabidopsis thaliana, partial (32%)
Length = 636
Score = 236 bits (602), Expect = 2e-62
Identities = 109/169 (64%), Positives = 137/169 (80%)
Frame = +2
Query: 330 VNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVV 389
V +LST +SI VD+LGRR LL+ G+QM + Q+++AIILGIK D+ + LSKGY++ VV
Sbjct: 2 VLLLSTFISIAIVDRLGRRPLLISGGIQMIICQVIVAIILGIKFGDNQE-LSKGYSLSVV 178
Query: 390 ILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHF 449
+ +C FV AF WSWGPLGW +PSE FPLE RSAGQS+TV VN+LFTF+IAQ FLS+LC F
Sbjct: 179 VAICLFVLAFGWSWGPLGWTVPSEIFPLEIRSAGQSITVAVNLLFTFIIAQTFLSLLCSF 358
Query: 450 KFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFM 498
KFGIFLFF+GW+ IM+IFV+ +PETK IPIEEM +WK+HWFWKR +
Sbjct: 359 KFGIFLFFAGWITIMTIFVVLFLPETKGIPIEEMA-IMWKKHWFWKRIL 502
>BG450455 similar to SP|Q07423|HEX6 Hexose carrier protein HEX6. [Castor
bean] {Ricinus communis}, partial (36%)
Length = 573
Score = 212 bits (540), Expect = 2e-55
Identities = 106/183 (57%), Positives = 139/183 (75%)
Frame = +2
Query: 9 SGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEA 68
S G + K+TPIVI+SC++AATGG++FGYD+G+SGGVTSM PFL+KFFP VY K +
Sbjct: 26 SNGRGYNGKMTPIVILSCMVAATGGIIFGYDIGISGGVTSMVPFLEKFFPDVYTKMKQDN 205
Query: 69 GLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNA 128
+ SNYCK+D+Q L FTSSLY+A L ++FFAS TR GR+ ++L+ G F+ G A
Sbjct: 206 KI-SNYCKFDSQLLTTFTSSLYIAGLLASFFASSITRAFGRKPSILVGGAAFLIGAALGG 382
Query: 129 AAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLV 188
AA N+ MLI+GR+LLG G+GFANQAVP++LS +A R RGA+NI FQL V IG+L NL+
Sbjct: 383 AALNIYMLILGRVLLGVGIGFANQAVPLYLSXMALPRYRGAINIGFQLCVGIGVLSXNLI 562
Query: 189 NYG 191
N+G
Sbjct: 563 NFG 571
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 209 bits (533), Expect = 2e-54
Identities = 147/513 (28%), Positives = 245/513 (47%), Gaps = 21/513 (4%)
Frame = +1
Query: 27 IMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDNQGLQLFT 86
++A+ ++ GYD+GV G A+Y K L K + +++
Sbjct: 124 MLASMTSILLGYDIGVMSGA------------AIYIKRDL---------KVSDGKIEVLL 240
Query: 87 SSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCG 146
+ + +L + A T+ +GRR T++ AG F G + N L+ GR + G G
Sbjct: 241 GIINIYSLIGSCLAGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYNFLMFGRFVAGVG 420
Query: 147 VGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLG 206
+G+A PV+ +E++P+ RG L ++ + GIL + NY +K+S GWR+ LG
Sbjct: 421 IGYALMIAPVYTAEVSPASSRGFLTSFPEVFINSGILLGYISNYAFSKLSLKLGWRMMLG 600
Query: 207 LAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAK 266
+ IP+++L VG + + ++P L+ RGRL + VL K +D+ E L L E + A
Sbjct: 601 VGAIPSVILAVGVLAMPESPRWLVMRGRLGDAIKVLNKT--SDSKEEAQLRLAEIKQAAG 774
Query: 267 EVKQAFRNLL--KGEKGGQ-----------------VIMAIGLEIFQQFTGINAIMFYAP 307
+ +++ K + G+ VI A+G+ FQQ +G++A++ Y+P
Sbjct: 775 IPEDCNDDVVEVKVKNTGEGVWKELFLYPTPAVRHIVIAALGIHFFQQASGVDAVVLYSP 954
Query: 308 VLFNTVGFKNDASLYSAVIT-GAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIA 366
+F G D L A I G V L +V+ + +D+ GRR LLL + M +S + +A
Sbjct: 955 TIFKKAGINGDTHLLIATIAVGFVKTLFILVATFMLDRYGRRPLLLTSVGGMVVSLLTLA 1134
Query: 367 IILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSV 426
+ L I DHS+ + V ++V+ F+ GP+ W+ SE FPL R+ G +
Sbjct: 1135VSLTI--IDHSNTKLNWAIGLSIATVLSYVATFSIGAGPITWVYSSEIFPLRLRAQGAAC 1308
Query: 427 TVCVNMLFTFVIAQAFLSMLCHFKF-GIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTE 485
V VN + + VI+ FLS+ G F F G I IF ++PET+ +EEM
Sbjct: 1309GVVVNRVTSGVISMTFLSLSKGITIGGAFFLFGGIATIGWIFFYIMLPETQGKTLEEMEA 1488
Query: 486 RVWKQHWFWKRFMEDDNEKVSNADYPKIKNNPN 518
K W++ + NA +++ N
Sbjct: 1489SFGK---IWRKSKNTKEAEKDNAQVAQVQLGTN 1578
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 184 bits (467), Expect = 7e-47
Identities = 118/354 (33%), Positives = 187/354 (52%), Gaps = 6/354 (1%)
Frame = +2
Query: 20 PIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDN 79
P V+ A GGL+FGYD GV G +++ FPAV +KT L+ +
Sbjct: 215 PYVLRLAFSAGIGGLLFGYDTGVISGALL---YIRDEFPAVEKKTWLQEAI--------- 358
Query: 80 QGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVG 139
S+ A+ + GR++++++A F+ G AAA N A LIVG
Sbjct: 359 ------VSTAIAGAIIGAAIGGWINDRFGRKVSIIVADTLFLLGSIILAAAPNPATLIVG 520
Query: 140 RILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGW 199
R+ +G GVG A+ A P+++SE +P+R+RGAL L +T G + L+N K G W
Sbjct: 521 RVFVGLGVGMASMASPLYISEASPTRVRGALVSLNSFLITGGQFLSYLINLAFTKAPGTW 700
Query: 200 GWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELC 259
W LG+A PA++ V + + ++P L +G+ EE K +LKKI ++ + E L
Sbjct: 701 RW--MLGVAAAPAVIQIVLMLSLPESPRWLYRKGKEEEAKVILKKIYEVEDYDNEIQALK 874
Query: 260 EASRVAKEVKQ----AFRNLLKGEKGGQVIMA-IGLEIFQQFTGINAIMFYAPVLFNTVG 314
E+ V E+K+ + L+K + + A +GL FQQFTGIN +M+Y+P + G
Sbjct: 875 ES--VEMELKETEKISIMQLVKTTSVRRGLYAGVGLAFFQQFTGINTVMYYSPSIVQLAG 1048
Query: 315 FKND-ASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAI 367
F + +L ++IT +N +I+SIYF+DK GR+ L L + + LS ++ +
Sbjct: 1049FASKRTALLLSLITSGLNAFGSILSIYFIDKTGRKKLALISLTGVVLSLTLLTV 1210
Score = 56.6 bits (135), Expect = 2e-08
Identities = 35/130 (26%), Positives = 63/130 (47%), Gaps = 8/130 (6%)
Frame = +2
Query: 375 DHSDDLSKG----YAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCV 430
DH +KG + ++ + ++ F+ G + W++ SE +PL R +
Sbjct: 1442 DHRAWYTKGCPSNFGWIAILALALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTT 1621
Query: 431 NMLFTFVIAQAFLSMLCHF-KFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEE---MTER 486
+ V++Q+FLS+ F+ F+ ++ FV+ VPETK +P+EE M E+
Sbjct: 1622 VWVSNLVVSQSFLSLTVAIGPAWTFMIFAIIAIVAIFFVIIFVPETKGVPMEEVESMLEK 1801
Query: 487 VWKQHWFWKR 496
+ Q FWK+
Sbjct: 1802 RFVQIKFWKK 1831
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 169 bits (429), Expect = 2e-42
Identities = 124/398 (31%), Positives = 202/398 (50%), Gaps = 5/398 (1%)
Frame = +3
Query: 91 LAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFA 150
+ A+ S A Y +GR+ +++IA I G + A++ + L +GR+L G GVG
Sbjct: 15 VGAIASGQIAEY----VGRKGSLMIASIPNIIGWLAISFAKDSSFLFMGRLLEGFGVGII 182
Query: 151 NQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLGLAGI 210
+ VPV+++EIAP +RG+L + QL+VTIGI+ A L+ N WR+ L +
Sbjct: 183 SYVVPVYIAEIAPENMRGSLGSVNQLSVTIGIMLAYLLGLFAN-------WRVLAILGIL 341
Query: 211 PALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTD-NIEPEFLELCEASRVAKEVK 269
P +L G + ++P L + G +EE + L+ +RG D +I E E+ +A VA K
Sbjct: 342 PCTVLIPGLFFIPESPRWLAKMGMMEEFETSLQVLRGFDTDISVEVHEIKKA--VASNGK 515
Query: 270 QA---FRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVI 326
+A F +L + + + IGL + QQ +GIN ++FY+ +F G +S + V
Sbjct: 516 RATIRFADLQRKRYWFPLSVGIGLLVLQQLSGINGVLFYSTSIFANAGI--SSSNAATVG 689
Query: 327 TGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIK-VTDHSDDLSKGYA 385
GA+ V++T V+ + VDK GRR+LL+ + M S +V++I ++ V +
Sbjct: 690 LGAIQVIATGVATWLVDKSGRRVLLIISSSLMTASLLVVSIAFYLEGVVEKDSQYFSILG 869
Query: 386 IFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSM 445
I V+ + V F+ GP+ WLI SE P+ + S N L ++I +
Sbjct: 870 IISVVGLVVMVIGFSLGLGPIPWLIMSEILPVNIKGLAGSTATMANWLVAWIITMTANLL 1049
Query: 446 LCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEM 483
L G FL ++ +F VPETK +EE+
Sbjct: 1050LTWSSGGTFLIYTVVAAFTVVFTSLWVPETKGRTLEEI 1163
>TC78543 similar to PIR|T14545|T14545 probable sugar transporter protein -
beet, partial (93%)
Length = 1653
Score = 165 bits (418), Expect = 3e-41
Identities = 134/472 (28%), Positives = 220/472 (46%), Gaps = 7/472 (1%)
Frame = +1
Query: 21 IVIISCIM-AATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDN 79
I +I+C++ A G + FG+ G + S + + GL +
Sbjct: 142 ISVIACVLIVALGPIQFGFTAGYTSPTQSA--------------IITDLGLSVSE----- 264
Query: 80 QGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVG 139
LF S + A+ + MGR+ +++IA I G + A + + L +G
Sbjct: 265 --FSLFGSLSNVGAMVGAIASGQIAEYMGRKGSLMIASIPNIIGWLMISFANDSSFLYMG 438
Query: 140 RILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGW 199
R+L G GVG + VPV+++EI+P +RG+L + QL+VT+GI+ A L+
Sbjct: 439 RLLEGFGVGIISYTVPVYIAEISPQNLRGSLVSVNQLSVTLGIMLAYLLGLFVE------ 600
Query: 200 GWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTD-NIEPEFLEL 258
WR L IP LL G + ++P L + G EE + L+ +RG + +I E E+
Sbjct: 601 -WRFLAILGIIPCTLLIPGLFFIPESPRWLAKMGMTEEFENSLQVLRGFETDISVEVNEI 777
Query: 259 CEASRVA-KEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGF-K 316
A A + F L + +++ IGL + QQ +GIN ++FY+ +F G
Sbjct: 778 KTAVASANRRTTVRFSELKQRRYWLPLMIGIGLLVLQQLSGINGVLFYSSTIFQNAGISS 957
Query: 317 NDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDH 376
+D + + GAV VL+T ++++ DK GRR+LL+ + M LS +V++I +K
Sbjct: 958 SDVATFG---VGAVQVLATTLTLWLADKSGRRLLLIVSSSAMTLSLLVVSISFYLKDYYI 1128
Query: 377 SDDLSKGYAIFVVILVC---TFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNML 433
S D S Y I ++ V V AF+ G + W+I SE P+ + S N
Sbjct: 1129SAD-SSLYGILSLLSVAGVVVMVIAFSLGMGAMPWIIMSEILPINIKGLAGSFATLANWF 1305
Query: 434 FTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTE 485
F+++I +L G F ++ FV VPETK +EE+ +
Sbjct: 1306FSWLITLTANLLLDWSSGGTFTIYTVVCAFTVGFVAIWVPETKGKTLEEIQQ 1461
>BI310242 similar to GP|347853|gb|A glucose transporter {Saccharum hybrid
cultivar H65-7052}, partial (36%)
Length = 519
Score = 163 bits (413), Expect = 1e-40
Identities = 72/107 (67%), Positives = 88/107 (81%)
Frame = -1
Query: 395 FVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIF 454
FV AF WSWGPLGW +PSE FPLE RSAGQS+TV VN+LFTF+IAQAFLS+LC FK+GIF
Sbjct: 519 FVLAFGWSWGPLGWTVPSEIFPLEIRSAGQSITVAVNLLFTFIIAQAFLSLLCFFKYGIF 340
Query: 455 LFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFMEDD 501
LFF+GW +M++FV +PETK IPIEEM+ + ++HWFWK + DD
Sbjct: 339 LFFAGWTALMTLFVFLFLPETKGIPIEEMS-ILLRKHWFWKMVLPDD 202
>AL368995 similar to GP|2104547|gb| AGAA.1 {Arabidopsis thaliana}, partial
(57%)
Length = 447
Score = 137 bits (345), Expect = 1e-32
Identities = 67/136 (49%), Positives = 95/136 (69%)
Frame = +1
Query: 5 GFATSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKT 64
G A ++ ++T VI++CI+AATGG +FGYDVG+SGGVTSM FL KFFP+VY++
Sbjct: 46 GVAKVRADQYNGRVTVHVILACIVAATGGSLFGYDVGISGGVTSMDDFLLKFFPSVYKQK 225
Query: 65 VLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGV 124
+ ++NYCKY+NQ L FTS LY++ L ++ AS TR GR++++++ G F+ G
Sbjct: 226 M--HAHENNYCKYNNQVLAAFTSVLYISGLVASLVASTITRKYGRKISIIVGGISFLIGS 399
Query: 125 AFNAAAQNLAMLIVGR 140
NAAA NL MLI+GR
Sbjct: 400 ILNAAAANLGMLIIGR 447
>TC83213 weakly similar to GP|16945177|emb|CAC69071. STP5 protein
{Arabidopsis thaliana}, partial (18%)
Length = 563
Score = 136 bits (342), Expect = 2e-32
Identities = 54/99 (54%), Positives = 77/99 (77%)
Frame = +1
Query: 401 WSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGW 460
WSWGPL WLIPSE FP++ R+ GQS+ V V + FV++Q FL+MLCH KFG F+F++ W
Sbjct: 1 WSWGPLTWLIPSEIFPVKIRTTGQSIAVAVQFIIIFVLSQTFLTMLCHMKFGAFVFYAFW 180
Query: 461 VLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFME 499
V++M++FV+F +PETK IP+E M +W +HWFW R+++
Sbjct: 181 VIVMTMFVIFFLPETKGIPLESM-YTIWGRHWFWSRYVK 294
>AL368310 weakly similar to GP|16945177|em STP5 protein {Arabidopsis
thaliana}, partial (22%)
Length = 477
Score = 130 bits (328), Expect = 9e-31
Identities = 69/124 (55%), Positives = 89/124 (71%), Gaps = 1/124 (0%)
Frame = +2
Query: 17 KITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSN-YC 75
K+T +II+CI+AA+GGL++GYD+GVSGGVT+M PFL+KFFP + RK A + N YC
Sbjct: 113 KLTLSIIITCIVAASGGLLYGYDLGVSGGVTTMVPFLQKFFPDILRKA---ASAEVNMYC 283
Query: 76 KYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAM 135
YD+Q L LFTSSLYLA L S+ AS T GRR ++I G FIAG A N ++N+ M
Sbjct: 284 VYDSQILTLFTSSLYLAGLVSSIAASXVTAAYGRRNVIIIGGALFIAGGAINGGSENIPM 463
Query: 136 LIVG 139
LI+G
Sbjct: 464 LILG 475
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 127 bits (319), Expect = 1e-29
Identities = 77/274 (28%), Positives = 136/274 (49%)
Frame = +3
Query: 93 ALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQ 152
AL + + +GRR + I G A +AA NL ++VGR+ +G G+G
Sbjct: 528 ALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMSAATNNLFGMLVGRLFVGTGLGLGPP 707
Query: 153 AVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPA 212
++++E++P+ +RG L Q+ GIL + + +IS GW WR+ ++ IPA
Sbjct: 708 VAALYVTEVSPAFVRGTYGALIQIATCFGILGSLFIGIPVKEIS-GW-WRVCFWVSTIPA 881
Query: 213 LLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAF 272
+L + ++P+ L ++GR E +A +++ G + +L + R F
Sbjct: 882 AILALAMDFCAESPHWLYKQGRTAEAEAEFERLLGVSEAKFAMSQLSKVDRGEDTDTVKF 1061
Query: 273 RNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNV 332
LL G V + L QQ +GINA+ +++ +F + G +D ++ V G N+
Sbjct: 1062SELLHGHHSKVVFIGSTLFALQQLSGINAVFYFSSTVFKSAGVPSD---FANVCIGVANL 1232
Query: 333 LSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIA 366
+I+S +DKLGR++LL + M +S I+ A
Sbjct: 1233TGSIISTGLMDKLGRKVLLFWSFFGMAISMIIQA 1334
>AW586674 weakly similar to SP|Q10710|STA_ Sugar carrier protein A. [Castor
bean] {Ricinus communis}, partial (21%)
Length = 531
Score = 121 bits (304), Expect = 6e-28
Identities = 63/119 (52%), Positives = 85/119 (70%), Gaps = 1/119 (0%)
Frame = +3
Query: 3 GGGFATSGGGE-FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVY 61
GGG G E ++ ++T VII+CI+AATGG +FGYDVG+SGGV SM FL+ FFPAVY
Sbjct: 180 GGGTVDKGRAEQYKGRVTVHVIIACIVAATGGSLFGYDVGISGGVASMDDFLQNFFPAVY 359
Query: 62 RKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFF 120
K LEA ++NYCKY+NQG+ FTS+LY++ L ++ A+ TR GRR +++I G F
Sbjct: 360 -KHKLEAH-ENNYCKYNNQGISAFTSTLYISGLVASIIAAPITRRYGRRTSIIIGGINF 530
>TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (55%)
Length = 949
Score = 121 bits (303), Expect = 7e-28
Identities = 85/282 (30%), Positives = 132/282 (46%), Gaps = 3/282 (1%)
Frame = +1
Query: 20 PIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDN 79
P ++ +A GGL+FGYD GV G ++K FP V L+
Sbjct: 163 PYILGLAAVAGIGGLLFGYDTGVISGALL---YIKDDFPQVRNSNFLQ------------ 297
Query: 80 QGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVG 139
+ S A+ F + GR+ L+A FI G AAA + +LI G
Sbjct: 298 ---ETIVSMAIAGAIVGAAFGGWLNDAYGRKKATLLADVIFILGAILMAAAPDPYVLIAG 468
Query: 140 RILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGW 199
R+L+G GVG A+ PV+++E+APS IRG+L L +T G + LVN ++ G W
Sbjct: 469 RLLVGLGVGIASVTAPVYIAEVAPSEIRGSLVSTNVLMITGGQFVSYLVNLVFTQVPGTW 648
Query: 200 GWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELC 259
W LG++G+PAL+ + + + ++P L + R E V+ KI +E E L
Sbjct: 649 RW--MLGVSGVPALIQFICMLFLPESPRWLFIKNRKNEAVDVISKIYDLSRLEDEIDFLT 822
Query: 260 EASRVAKEVKQA--FRNLLKGEKGGQVIM-AIGLEIFQQFTG 298
S ++ + F ++ + ++ + GL FQQFTG
Sbjct: 823 AQSEQERQRRSTIKFWHVFRSKETRLAFLDGGGLLAFQQFTG 948
>TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar transporter
{Prunus armeniaca}, partial (59%)
Length = 1074
Score = 117 bits (293), Expect = 1e-26
Identities = 77/286 (26%), Positives = 148/286 (50%), Gaps = 3/286 (1%)
Frame = +1
Query: 201 WRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCE 260
WR G+A +P++LL +G + ++P L ++G++ E + +K + G + + +L
Sbjct: 19 WRTMFGIAVVPSILLALGMAISPESPRWLFQQGKIAEAEKAIKTLYGKERVATVMYDLRT 198
Query: 261 ASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDAS 320
AS+ + E + + +L V + L +FQQ GINA+++Y+ +F + G +D +
Sbjct: 199 ASQGSSEPEAGWFDLFSSRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGIASDVA 378
Query: 321 LYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDDL 380
++ + GA NV+ T ++ +DK GR+ LL+ + M S +++++ KV L
Sbjct: 379 --ASALVGASNVIGTAIASSLMDKQGRKSLLITSFSGMAASMLLLSLSFTWKV------L 534
Query: 381 SKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQ 440
+ V+ +V +F+ GP+ L+ E F R+ S+++ + + FVI
Sbjct: 535 APYSGTLAVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVSLSLGTHWISNFVIGL 714
Query: 441 AFLSMLCHFKFGIFLFFSGWVLIMSIFVLFL---VPETKNIPIEEM 483
FLS++ KFGI + G+ + + VL++ V ETK +EE+
Sbjct: 715 YFLSVV--NKFGISSVYLGFSAVCVLAVLYIAGNVVETKGRSLEEI 846
>BQ751130 weakly similar to GP|15289796|dbj putative monosaccharide
transporter 3 {Oryza sativa (japonica cultivar-group)},
partial (7%)
Length = 795
Score = 105 bits (262), Expect = 4e-23
Identities = 72/256 (28%), Positives = 122/256 (47%), Gaps = 3/256 (1%)
Frame = +3
Query: 15 EAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNY 74
EA +T + C A+ GG+ FGYD G GV + F++ G +
Sbjct: 18 EAPVTWKAYVICAFASFGGIFFGYDSGYINGVLGANQFIEHV-----------EGPGAEA 164
Query: 75 CKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVA---FNAAAQ 131
+Q L S L A + +GR+ T++I ++ GV F
Sbjct: 165 ISESHQSL--IVSILSCGTFFGALIAGDVSDWIGRKWTVIIGCGIYMIGVLIQMFTGMGA 338
Query: 132 NLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYG 191
L ++ GR++ G GVGF + V +++SEI P ++RGAL +Q +TIG+L A V Y
Sbjct: 339 PLGAIVAGRLIAGIGVGFESAIVILYMSEICPKKVRGALVAGYQFCITIGLLLAACVVYA 518
Query: 192 TNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNI 251
T +R+ + + AL+L G +++ ++P +++GR+ + A L ++RG
Sbjct: 519 TKDRGDTGVYRIPIAIQFPWALILGGGLLLLPESPRFFVKKGRIADATAALSRLRGQPK- 695
Query: 252 EPEFLELCEASRVAKE 267
E E++++ A VA E
Sbjct: 696 ESEYIQVELAEIVANE 743
>BQ136503 weakly similar to PIR|T05156|T051 probable glucose transport
protein F18E5.100 - Arabidopsis thaliana, partial (15%)
Length = 490
Score = 100 bits (248), Expect = 2e-21
Identities = 43/85 (50%), Positives = 63/85 (73%)
Frame = +3
Query: 424 QSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEM 483
QS+TV VNM TF IAQ F MLCHFKFG+F+FF +V++M+ F+ PETK +P+EEM
Sbjct: 9 QSITVAVNMTSTFFIAQLFTEMLCHFKFGLFIFFGCFVILMTFFIYKFFPETKGVPLEEM 188
Query: 484 TERVWKQHWFWKRFMEDDNEKVSNA 508
VW++H FW +F+E +++K+ ++
Sbjct: 189 -HMVWRKHPFWGKFLEAEDQKIQSS 260
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,322,479
Number of Sequences: 36976
Number of extensions: 214205
Number of successful extensions: 1947
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 1852
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1883
length of query: 521
length of database: 9,014,727
effective HSP length: 100
effective length of query: 421
effective length of database: 5,317,127
effective search space: 2238510467
effective search space used: 2238510467
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC145222.4


