

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.11 + phase: 0 /pseudo
(394 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
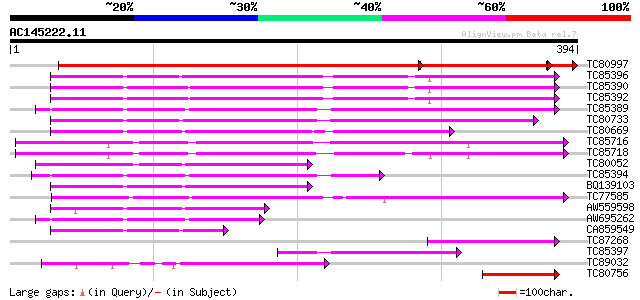
Score E
Sequences producing significant alignments: (bits) Value
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 499 0.0
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 185 3e-47
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 179 2e-45
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 178 3e-45
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 159 1e-39
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 159 1e-39
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 135 3e-32
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 132 2e-31
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 130 9e-31
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 110 1e-24
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 107 6e-24
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 96 3e-20
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 95 5e-20
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 75 3e-14
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 67 9e-12
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 64 1e-10
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 62 3e-10
TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated... 60 1e-09
TC89032 similar to GP|15293149|gb|AAK93685.1 putative HSP protei... 59 4e-09
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 54 1e-07
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 499 bits (1286), Expect(3) = 0.0
Identities = 253/253 (100%), Positives = 253/253 (100%)
Frame = +3
Query: 35 GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFN 94
GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFN
Sbjct: 3 GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFN 182
Query: 95 MKRLIGRVDTDPVVHASKNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLVELR 154
MKRLIGRVDTDPVVHASKNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLVELR
Sbjct: 183 MKRLIGRVDTDPVVHASKNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLVELR 362
Query: 155 LMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQQQQ 214
LMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQQQQ
Sbjct: 363 LMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQQQQ 542
Query: 215 KASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGSTIGGEDLLQNMMRHL 274
KASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGSTIGGEDLLQNMMRHL
Sbjct: 543 KASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGSTIGGEDLLQNMMRHL 722
Query: 275 LPDSENIFKRHGV 287
LPDSENIFKRHGV
Sbjct: 723 LPDSENIFKRHGV 761
Score = 179 bits (453), Expect(3) = 0.0
Identities = 90/92 (97%), Positives = 91/92 (98%)
Frame = +1
Query: 286 GVEEIKSMALLRVATQDAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKC 345
G EEIKSMALLRVATQDAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKC
Sbjct: 757 GFEEIKSMALLRVATQDAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKC 936
Query: 346 ESLIIQCLHDAKVEVGDINDVILVGGCSYIPQ 377
ESLIIQCLHDAKVEVGDINDVILVGGCSYIP+
Sbjct: 937 ESLIIQCLHDAKVEVGDINDVILVGGCSYIPR 1032
Score = 43.5 bits (101), Expect(3) = 0.0
Identities = 19/21 (90%), Positives = 20/21 (94%)
Frame = +2
Query: 374 YIPQGWRILLPTYAKLQKFIK 394
+I QGWRILLPTYAKLQKFIK
Sbjct: 1019 HISQGWRILLPTYAKLQKFIK 1081
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 185 bits (469), Expect = 3e-47
Identities = 122/361 (33%), Positives = 196/361 (53%), Gaps = 7/361 (1%)
Frame = +2
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 131 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 304
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF V G +P I + EE+ +
Sbjct: 305 TNTVFDAKRLIGRRISDASVQSDMKLWPFKV-IAGPGEKPMIGVNYKGEEKLFASEEISS 481
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 482 MVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 661
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 662 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 823
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 824 DNRMVTHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFE 991
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 992 GVDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQL 1171
Query: 382 L 382
L
Sbjct: 1172L 1174
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 179 bits (454), Expect = 2e-45
Identities = 119/361 (32%), Positives = 195/361 (53%), Gaps = 7/361 (1%)
Frame = +1
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 160 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 333
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D V + L PF V +P I + + EE+ +
Sbjct: 334 TNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPAD-KPMIVVNYKAEEKQFSAEEISS 510
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L++++ + E +L I+N V+TVP F+ Q + A ++GL+VLR++ EPTA A+
Sbjct: 511 MVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIINEPTAAAI 690
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 691 AYGLDKKATST------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 852
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 853 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFE 1020
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 1021GIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQL 1200
Query: 382 L 382
L
Sbjct: 1201L 1203
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 178 bits (452), Expect = 3e-45
Identities = 117/361 (32%), Positives = 196/361 (53%), Gaps = 7/361 (1%)
Frame = +1
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 154 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 327
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF V +P I + + EE+ +
Sbjct: 328 TNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPAD-KPMIVVNYKAEEKQFSAEEISS 504
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L++++ + E +L I+N V+TVP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 505 MVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAI 684
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 685 AYGLDKKATST------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 846
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ +R A + A LS+ + +++D L
Sbjct: 847 DNRMVNHFVQE----FKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFE 1014
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 1015GIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQL 1194
Query: 382 L 382
L
Sbjct: 1195L 1197
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 159 bits (403), Expect = 1e-39
Identities = 112/368 (30%), Positives = 200/368 (53%), Gaps = 4/368 (1%)
Frame = +2
Query: 19 EEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTS 78
EE ++ L + IGID+GT+ V V+ VE++ N + ++ S+V+F D+ G +
Sbjct: 134 EEGTTKLGTV-IGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAA 310
Query: 79 QFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNN-V 136
+ + + TIF++KRLIGR D V L P+ + D +P+I V +
Sbjct: 311 K--NLAAVNPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKD--GKPYIQVRVKDGE 478
Query: 137 WRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVL 196
+ +PEEV AM L +++ E L + IR+ V+TVP F+ Q + A +AGL+V
Sbjct: 479 TKVFSPEEVSAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVA 658
Query: 197 RLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKAL 256
R++ EPTA A+ YG ++ G EK L+F++G G DV++ GV ++ +
Sbjct: 659 RIINEPTAAAIAYGLDKK---------GGEKNILVFDLGGGTFDVSILTIDNGVFEVLST 811
Query: 257 AGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQV 315
G T +GGED Q +M + + + + ++ +++ LR ++ A LS+Q V+V
Sbjct: 812 NGDTHLGGEDFDQRIMEYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRV 991
Query: 316 DVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSY 374
+++ L + + + + RA FEE+N ++F K + + + DA ++ I++++LVGG +
Sbjct: 992 EIESLFDGVDFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTR 1171
Query: 375 IPQGWRIL 382
IP+ ++L
Sbjct: 1172IPKVQQLL 1195
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 159 bits (403), Expect = 1e-39
Identities = 104/342 (30%), Positives = 183/342 (53%), Gaps = 3/342 (0%)
Frame = +2
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V + ++E++ N + + S+V F D G ++ ++ M
Sbjct: 188 AIGIDLGTTYSCVGRYANDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAK--NQVAMNP 361
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V A K+ PF V +D G +P I ++ TPEE+ A
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKV--IDKGGKPNIEVEFKGENKTFTPEEISA 535
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M LV++R E +L + N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 536 MVLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAI 715
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ + E+ LIF++G G DV++ G+ ++K+ AG T +GGED
Sbjct: 716 AYGLDKK--------AEGERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDF 871
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
++ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 872 DNRLVNHFVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDF 1051
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVI 367
+ RA FEE+ +++F + + L DAK++ +++++
Sbjct: 1052YTSITRARFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 135 bits (340), Expect = 3e-32
Identities = 94/283 (33%), Positives = 149/283 (52%), Gaps = 2/283 (0%)
Frame = +2
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 119 AIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 292
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V L PF V +P I + EE+ +
Sbjct: 293 QNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVP-GPAEKPMIVVNYKGEEKKFAAEEISS 469
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E L P++N V+TVP F+ Q + A A++GL+VLR++ EPTA A+
Sbjct: 470 MVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAI 649
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS++ E+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 650 AYG-LDKKASRK-----GEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 811
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLST 309
M+ H + + K+ +++ LR A + A LS+
Sbjct: 812 DNRMVNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSS 940
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 132 bits (332), Expect = 2e-31
Identities = 101/393 (25%), Positives = 185/393 (46%), Gaps = 9/393 (2%)
Frame = +1
Query: 5 EPAYTVASDSETTGEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSF 64
+P+Y + S P IGID+GT+ V++ G ++++N + S
Sbjct: 172 KPSYAAQKWASLARPFSSRPAGSDVIGIDLGTTNSCVSLMEGKNPKVIENSEGARTTPSV 351
Query: 65 VTF--KDEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTL 121
V F K E G + + + +T+F KRLIGR DP K +P+ +
Sbjct: 352 VAFNQKGELLVGTPAKRQAVTNPT---NTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKA 522
Query: 122 DIGVRPFIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQL 181
G A V + +P ++ A L +++ E +L + I V+TVP F+ Q
Sbjct: 523 PNG-----DAWVEINKQQYSPSQIGAFVLTKMKETAEAYLGKTISKAVVTVPAYFNDAQR 687
Query: 182 TRLERACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDV 241
+ A +AGL V R++ EPTA AL YG + E + +F++G G DV
Sbjct: 688 QATKDAGRIAGLEVKRIINEPTAAALSYGM-----------NNKEGLIAVFDLGGGTFDV 834
Query: 242 AVTATAGGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVAT 300
++ + GV ++KA G T +GGED ++ L+ + + ++ ++ LR A
Sbjct: 835 SILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVSEFKRTDSIDLAKDKLALQRLREAA 1014
Query: 301 QDAIHQLSTQSSVQVDV-----DLGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHD 355
+ A +LS+ S ++++ D + + + R++FE + + E+ ++ CL D
Sbjct: 1015EKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCKSCLKD 1194
Query: 356 AKVEVGDINDVILVGGCSYIPQGWRILLPTYAK 388
A + + D+++V+LVGG + +P+ ++ + K
Sbjct: 1195ANISIKDVDEVLLVGGMTRVPKVQEVVSEIFGK 1293
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 130 bits (327), Expect = 9e-31
Identities = 108/397 (27%), Positives = 187/397 (46%), Gaps = 13/397 (3%)
Frame = +2
Query: 5 EPAYTVASDSETTGEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSF 64
+PAY + S + S IGID+GT+ V+V G ++++N + S
Sbjct: 188 KPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSV 367
Query: 65 VTF--KDEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTL 121
V F K E G + + + +TI KRLIGR DP K +P+ +
Sbjct: 368 VAFTQKGELLVGTPAKRQAVTNPE---NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKA 538
Query: 122 DIGVRPFIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQL 181
G A V + +P ++ A L +++ E +L + + V+TVP F+ Q
Sbjct: 539 PNG-----DAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQR 703
Query: 182 TRLERACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDV 241
+ A +AGL VLR++ EPTA AL YG + E + +F++G G DV
Sbjct: 704 QATKDAGRIAGLEVLRIINEPTAAALSYGMNK------------EGLIAVFDLGGGTFDV 847
Query: 242 AVTATAGGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIK----SMALL 296
++ + GV ++KA G T +GGED ++ L+ N FKR ++ ++ L
Sbjct: 848 SILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV----NEFKRSESIDLSKDKLALQRL 1015
Query: 297 RVATQDAIHQLSTQSSVQVDV-----DLGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQ 351
R A + A +LS+ S ++++ D + + + R++FE + + E+ ++
Sbjct: 1016REAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQS 1195
Query: 352 CLHDAKVEVGDINDVILVGGCSYIPQGWRILLPTYAK 388
CL DA + DI++V+LVGG + +P+ ++ + K
Sbjct: 1196CLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGK 1306
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 110 bits (274), Expect = 1e-24
Identities = 72/194 (37%), Positives = 109/194 (56%), Gaps = 2/194 (1%)
Frame = +3
Query: 19 EEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKD-EAPSGGVT 77
+E S ++AIGID+GT+ VAV E++++ N ++ SFV FKD E G
Sbjct: 75 KEMSGRGKKVAIGIDLGTTYSCVAVCKNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAA 254
Query: 78 SQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNV 136
+ + +TIF+ KRLIGR +DP+V + L PF V D+ +P I N+
Sbjct: 255 FNIAASNPT---NTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIG-DLNDKPMIVVNYNDE 422
Query: 137 WRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVL 196
+ EE+ +M LV++R + ET L + +VV+TVP F+ Q A A+AGL+V+
Sbjct: 423 EKHFAAEEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVM 602
Query: 197 RLMPEPTAVALLYG 210
R++ EPTA A+ YG
Sbjct: 603 RIINEPTAAAIAYG 644
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 107 bits (268), Expect = 6e-24
Identities = 78/247 (31%), Positives = 131/247 (52%), Gaps = 2/247 (0%)
Frame = +1
Query: 16 TTGEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGG 75
+ +E+++ L + IGID+GT+ V V+ VE++ N + ++ S+V+F D+ G
Sbjct: 259 SNAKEEATKLGTV-IGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIG 435
Query: 76 VTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVN 134
++ + + TIF++KRLIGR D V L P+ + D +P+I V
Sbjct: 436 EAAK--NLAAVNPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNRD--GKPYIQVRVK 603
Query: 135 N-VWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGL 193
+ + +PEE+ AM L +++ E L + IR+ V+TVP F+ Q + A +AGL
Sbjct: 604 DGETKVFSPEEISAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGL 783
Query: 194 HVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQI 253
+V R++ EPTA A+ YG ++ G EK L+F++G G DV++ GV ++
Sbjct: 784 NVARIINEPTAAAIAYGLDKK---------GGEKNILVFDLGGGTFDVSILTIDNGVFEV 936
Query: 254 KALAGST 260
A G T
Sbjct: 937 LATNGDT 957
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 95.5 bits (236), Expect = 3e-20
Identities = 62/183 (33%), Positives = 99/183 (53%), Gaps = 1/183 (0%)
Frame = +2
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 80 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 253
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF + + +P I + EE+ +
Sbjct: 254 INTVFDAKRLIGRRFSDASVQSDMKLWPFKIIS-GPAEKPLIGVNYKGEDKEFAAEEISS 430
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 431 MVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 610
Query: 208 LYG 210
YG
Sbjct: 611 AYG 619
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 94.7 bits (234), Expect = 5e-20
Identities = 90/364 (24%), Positives = 163/364 (44%), Gaps = 5/364 (1%)
Frame = +1
Query: 30 IGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFG 89
+G D G C VAV ++++ N +++ + V F D+ G S M
Sbjct: 247 VGFDFGNESCIVAVARQRGIDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTM--MNPK 420
Query: 90 DTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAM 148
++I +KRLIG+ DP + K+LPF V G P I A R T +V M
Sbjct: 421 NSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGY-PLIHARYLGESREFTATQVFGM 597
Query: 149 FLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALL 208
L L+ + + +L + + + +PV F+ Q + A +AGLH L L+ E TA AL
Sbjct: 598 MLSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALA 777
Query: 209 YGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGS---TIGGED 265
YG + + +E + + F + G+ + V Q+ L+ S ++GG D
Sbjct: 778 YGIYKTDLPE------NEWLNVAF-VDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRD 936
Query: 266 LLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLK 324
+ + H + +K + ++ LR A + LS ++++ L +
Sbjct: 937 FDEALFHHFAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKD 1116
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRILLP 384
+ + R +FE+++ + E+ + + + L +A + V +I+ V +VG S +P +IL
Sbjct: 1117VRGFIKRDDFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTE 1296
Query: 385 TYAK 388
+ K
Sbjct: 1297FFKK 1308
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 75.5 bits (184), Expect = 3e-14
Identities = 51/155 (32%), Positives = 82/155 (52%), Gaps = 3/155 (1%)
Frame = +1
Query: 29 AIGIDIGTSQCSVAVW--NGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEM 86
A+GID+GT+ VAVW + VE++ N K+ SFV F D G ++ + +
Sbjct: 55 AVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAAK--DQAAI 228
Query: 87 LFGDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEV 145
+T+F+ KRLIGR +DPVV L PF V + + +P I+ + EE+
Sbjct: 229 NPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVS-GVNDKPMISLKFKGQEKLLCAEEI 405
Query: 146 LAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQ 180
++ L +R + E +L+ P++N +TVP F+ Q
Sbjct: 406 SSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQ 510
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 67.4 bits (163), Expect = 9e-12
Identities = 52/161 (32%), Positives = 86/161 (53%), Gaps = 2/161 (1%)
Frame = +2
Query: 19 EEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTS 78
EE ++ L + IGID+GT+ V V+ VE++ N + ++ S+V+F D+ G +
Sbjct: 107 EEGTTKLGTV-IGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAA 283
Query: 79 QFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALV-NNV 136
+ + + TIF++KRLIGR D V K +P+ + D +P+I V +
Sbjct: 284 K--NLAAVNPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKD--GKPYIQVRVKDGE 451
Query: 137 WRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFS 177
+ +PEEV AM L +++ E L + IR+ V+TVP S
Sbjct: 452 TKVFSPEEVSAMILTKMKETAEAFLGKTIRDAVVTVPALLS 574
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 63.5 bits (153), Expect = 1e-10
Identities = 41/125 (32%), Positives = 65/125 (51%), Gaps = 1/125 (0%)
Frame = +3
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 165 AVGIDLGTTYSCVGVWQNDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 338
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D V + K+ PF V +D +P+I + TPEE+ +
Sbjct: 339 YNTVFDAKRLIGRRFADQEVQSDMKHWPFKV--IDKAAKPYIQVEYKGETKQFTPEEISS 512
Query: 148 MFLVE 152
M L +
Sbjct: 513 MVLTK 527
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 62.4 bits (150), Expect = 3e-10
Identities = 31/93 (33%), Positives = 56/93 (59%), Gaps = 1/93 (1%)
Frame = +2
Query: 291 KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLI 349
+++ LR A + A LS+ + +++D L + + RA FEE+N ++F KC +
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 350 IQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLL 280
>TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated protein 78
{Lycopersicon esculentum}, partial (57%)
Length = 648
Score = 60.5 bits (145), Expect = 1e-09
Identities = 39/129 (30%), Positives = 68/129 (52%), Gaps = 1/129 (0%)
Frame = +1
Query: 187 ACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTAT 246
A +AGL+V R++ EPTA A+ YG ++ G EK L+F++G G DV++
Sbjct: 289 AGVIAGLNVARIINEPTAAAIAYGLDKK---------GGEKNILVFDLGGGTFDVSILTI 441
Query: 247 AGGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIH 305
GV ++ A G T +GGED Q + + + + + ++ +++ LR ++ A
Sbjct: 442 DNGVFEVLATNGDTHLGGEDFDQRIREYFIKLIKKKHSKDISKDNRALGKLRKESERAKR 621
Query: 306 QLSTQSSVQ 314
LS+Q V+
Sbjct: 622 ALSSQHQVR 648
Score = 35.0 bits (79), Expect = 0.050
Identities = 28/101 (27%), Positives = 46/101 (44%), Gaps = 11/101 (10%)
Frame = +1
Query: 36 TSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFNM 95
T+ V V+ VE++ N + ++ S+V+F D+ G ++ + + TIF++
Sbjct: 1 TTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAK--NLAAVNPERTIFDV 174
Query: 96 KRLIGRVDTDPVVHASKNL-----------PFLVQTLDIGV 125
KRLIGR D V L P++ T D GV
Sbjct: 175 KRLIGRKFEDKEVQRDMKLVPYKIVNRDGKPYIQATKDAGV 297
>TC89032 similar to GP|15293149|gb|AAK93685.1 putative HSP protein
{Arabidopsis thaliana}, partial (26%)
Length = 900
Score = 58.5 bits (140), Expect = 4e-09
Identities = 54/214 (25%), Positives = 96/214 (44%), Gaps = 14/214 (6%)
Frame = +1
Query: 23 SPLPEIAIGIDIGTSQCSVAVWN----GSEVELLKNKRNQKLMKSFVTFKDE-----APS 73
SP +D+G+ VAV N S + + N+ +++ V+F D +
Sbjct: 136 SPSHSAVFSVDLGSESLKVAVVNLKPGQSPISVAINEMSKRKSPVLVSFHDGDRLLGEEA 315
Query: 74 GGVTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASK-----NLPFLVQTLDIGVRPF 128
G+ +++ + M+ LIG+ P A K LPF + + R
Sbjct: 316 AGLVARYPQK-------VYSQMRDLIGK----PYASAKKFLDSMYLPF--EAKENSSRGT 456
Query: 129 IAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERAC 188
++ +V+ +PEE+ AM L + E H K PI++ V+ VP F + + L +A
Sbjct: 457 VSFVVDENGTEYSPEELTAMILTYAANLAEFHSKIPIKDAVIAVPPYFGQAERRGLLQAA 636
Query: 189 AMAGLHVLRLMPEPTAVALLYGQQQQKASQETMG 222
+AG++VL L+ E + AL YG + +++ G
Sbjct: 637 ELAGINVLSLINEYSGAALQYGIDKDFSNESRHG 738
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 53.5 bits (127), Expect = 1e-07
Identities = 22/54 (40%), Positives = 38/54 (69%)
Frame = +3
Query: 329 VDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ +++V+LVGG + IP+ ++L
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLL 164
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,249,337
Number of Sequences: 36976
Number of extensions: 122272
Number of successful extensions: 628
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 587
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 592
length of query: 394
length of database: 9,014,727
effective HSP length: 98
effective length of query: 296
effective length of database: 5,391,079
effective search space: 1595759384
effective search space used: 1595759384
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC145222.11


