

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
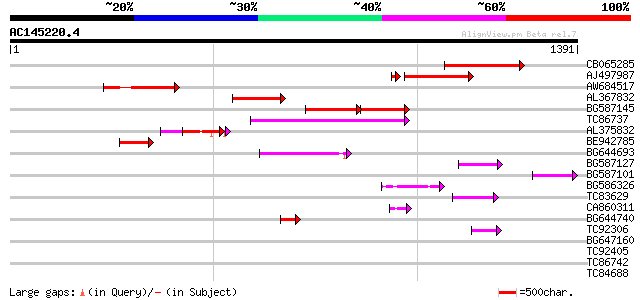
Query= AC145220.4 - phase: 0 /pseudo
(1391 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 409 e-114
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 298 1e-80
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 260 3e-69
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 244 2e-64
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 152 4e-59
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 179 7e-45
AL375832 141 2e-33
BE942785 137 4e-32
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 83 6e-16
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 67 6e-11
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 60 8e-09
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 51 4e-06
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 50 8e-06
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 49 1e-05
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 49 2e-05
TC92306 49 2e-05
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 37 0.070
TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~un... 36 0.12
TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG00... 35 0.26
TC84688 similar to GP|10241605|emb|CAC09581. protein kinase {Fag... 33 0.77
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 409 bits (1052), Expect = e-114
Identities = 195/197 (98%), Positives = 196/197 (98%)
Frame = -2
Query: 1066 KSKDCEEPLIGEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNN 1125
KSKDCEEPLIGEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNN
Sbjct: 591 KSKDCEEPLIGEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNN 412
Query: 1126 MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYF 1185
MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYF
Sbjct: 411 MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYF 232
Query: 1186 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFAIENVIDQTGE 1245
TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFAI NVIDQTGE
Sbjct: 231 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFAIGNVIDQTGE 52
Query: 1246 NVVDYKPWYYDIKQFLL 1262
NVVDYKPWYYDIKQFL+
Sbjct: 51 NVVDYKPWYYDIKQFLV 1
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 298 bits (763), Expect = 1e-80
Identities = 132/167 (79%), Positives = 156/167 (93%)
Frame = -2
Query: 970 KTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKNARWQMLLSEYDIVFKT 1029
KTCCALAWAAKRLRHY++NHTTWL+S+MDPIKYIFEK A+TG+ ARWQMLLSEYDI +++
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1030 QKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIGEGPDPNSKWGLVF 1089
QKAIKGSILADHLA+QPL+DY+PI+FDFPDEEIMYLK KDC+EPL GEGPDP+S WGL+F
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 1090 DGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGI 1136
DGAVN YG GIGAV+++P+G +IPFTAR+ F+CTNN+AEYEACI GI
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
Score = 48.1 bits (113), Expect = 2e-05
Identities = 20/24 (83%), Positives = 23/24 (95%)
Frame = +2
Query: 936 CVLGQQDETGKKEHAIYYLSKKFT 959
CVLGQQDETG+KEHAIYYLS+K +
Sbjct: 38 CVLGQQDETGRKEHAIYYLSRKLS 109
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 488
Score = 260 bits (664), Expect = 3e-69
Identities = 133/187 (71%), Positives = 146/187 (77%)
Frame = +1
Query: 231 VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNGKLVTVHGEEAYLVSQLSSF 290
+TFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 291 SCIEAGSAEGTAFQGLTIEGAEPKKAGAAMASLKDAQRVIQEGQTAGWGKVIQLHENKRK 350
SAEGTAFQGL++EGAEPKK GAAMASLKDAQ+ +QEGQ A WGK+IQL ENKRK
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 351 EGLGFSPSSRVSSGVFHSAGFVNTITEEATGSGLRPIFVIPGGIARDWDAIDIPSIMHVS 410
EGL FSP+S VS+G FHSAGFVNT+ EE RP+FVIPGGIA+DWDA+D+PSIMHVS
Sbjct: 286 EGLRFSPTSGVSTGTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIMHVS 465
Query: 411 E*CVLLL 417
*CVLLL
Sbjct: 466 X*CVLLL 486
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 244 bits (622), Expect = 2e-64
Identities = 119/128 (92%), Positives = 123/128 (95%)
Frame = -1
Query: 548 QEKKAIQPHQEEIELINIGIEENKREIKIGAALEEGVKKKIIQLLREYPDIFAWSYEDMP 607
QE+KAIQPHQEEIELIN+G EENKREIK+GAALEEGVK+KI QLLREY DIFA SYEDMP
Sbjct: 384 QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMP 205
Query: 608 GLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 667
GLDPKIVEHRIPTKPECPPVR KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI
Sbjct: 204 GLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 25
Query: 668 VPVPKKDG 675
VPVPKKDG
Sbjct: 24 VPVPKKDG 1
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 152 bits (384), Expect(2) = 4e-59
Identities = 70/136 (51%), Positives = 101/136 (73%)
Frame = +2
Query: 727 MSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVK 786
M P+D EKT+FIT GT+CYKVMPFGL NAG+TYQR + +F D + +EVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 787 STDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIR 846
S H+ +L + F+ L +Y ++LNP KCTFGV SG+ LG+IV+Q+GIEV+P ++ AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 847 EMPAPRTEKQVRGFLG 862
++P+P+ ++V+ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 95.9 bits (237), Expect(2) = 4e-59
Identities = 48/120 (40%), Positives = 74/120 (61%)
Frame = +3
Query: 862 GRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQEAFDSIKNYLLKPPILVPPVEGR 921
GR+ ++RFIS T C P +KLL N+ VW+++C+EAF+ +K YL PP+L P G
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 922 PLIMYLVVFDESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKR 981
L +Y+ + ++ VL ++D +K I+Y SK+ TD ETRY LEK A+ +A++
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQK--PIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 179 bits (454), Expect = 7e-45
Identities = 122/398 (30%), Positives = 199/398 (49%), Gaps = 7/398 (1%)
Frame = +1
Query: 591 LLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPEC----PPVRQ-KLRRTHPDMALKIKSE 645
+L E+PD+F +++H IP P+ PP+ L L +K
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 646 VQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDMLVD 705
++ +D GF+ A ++ V K G +R CVD+R LN + KD +PLP I +
Sbjct: 523 LEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLR 699
Query: 706 NTAQPKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMT 765
A + F+ +D + ++++++ ED+EKT+F T +G F + V PFGL A AT+QR +
Sbjct: 700 RVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYIN 879
Query: 766 TLFHDMIHKEVEVYVDDMIVKST-DEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGK 824
H+ + V Y+DD+++ +T ++ H + ++ RL L L+P KC F V + K
Sbjct: 880 KTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVK 1059
Query: 825 LLGFIVSQ-KGIEVDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFK 883
+GFI++ KG+ DP K+ AIR+ P + K R FLG NY FI + P+ +
Sbjct: 1060YVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTR 1239
Query: 884 LLRKNQPIVWNDECQEAFDSIKNYLLKPPILVPPVEGRPLIMYLVVFDESMGCVLGQQDE 943
L RK+ P W E + AF +K + P+L + ++G VL Q+D
Sbjct: 1240LTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDG 1419
Query: 944 TGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKR 981
TG H + + S++ + E Y + +K A+ WA R
Sbjct: 1420TG-AAHPVAFHSQRLSPAEYNYPIHDKELLAV-WACLR 1527
>AL375832
Length = 535
Score = 141 bits (356), Expect = 2e-33
Identities = 78/109 (71%), Positives = 83/109 (75%), Gaps = 6/109 (5%)
Frame = +1
Query: 423 KITILSLCPKRK*YFL*GRVCFGIAVDE*KLSFLSRLLFFLSAFFLGNNGNARKNQNICI 482
KITILSLCPKR+*YFL*GRVCFGIAV K+ FFL F NGNARKN N+CI
Sbjct: 160 KITILSLCPKRE*YFL*GRVCFGIAV**IKIVVSFTTAFFLVPFSW-KNGNARKNLNVCI 336
Query: 483 FL*ISKTRMKKK------NSF*NHPIIICRLNHNKPIEHSNPMPLPNFE 525
FL ISKT MKKK NSF*NHPIIICRLNHNKP+EHSN MPLPNF+
Sbjct: 337 FLLISKTCMKKKKYKKQKNSF*NHPIIICRLNHNKPVEHSNSMPLPNFD 483
Score = 101 bits (252), Expect = 2e-21
Identities = 80/178 (44%), Positives = 88/178 (48%), Gaps = 7/178 (3%)
Frame = +3
Query: 371 FVNTITEEATGSGLRPIFVIPGGIARDWDAIDIPSIMHVSE*CVLLL*YFVFKITILSLC 430
FVN I+EEATGSGLRP FV PGGIA DWDAIDIPSIMHVSE*CVLLL* FVFK
Sbjct: 3 FVNAISEEATGSGLRPAFVTPGGIASDWDAIDIPSIMHVSE*CVLLL*RFVFKNNNPLAL 182
Query: 431 PKRK*YFL*GRVCFGIAVDE*KLSFLSRLLFFLSAFFLGNNGNARKNQNICIFL*ISKTR 490
PK + F F K FF SAFFL +K FL K
Sbjct: 183 PKARVIFSVRACLFRHCRLMNKNCRFFHDCFFFSAFFL-EKW*CQKKPKRLYFLINFKNL 359
Query: 491 MKKKNSF*NHPIIICRLNHNKPIEHSNPMPLPNFE------FPVYEAEDEE-SDIPYE 541
+KK + + NH +E F FPVYE + E DIPYE
Sbjct: 360 HEKKKIQKTKKLFLKSSNHYMQVEP**TR*TQ*FYASSKL*FPVYEPKMREGDDIPYE 533
>BE942785
Length = 460
Score = 137 bits (344), Expect = 4e-32
Identities = 64/82 (78%), Positives = 73/82 (88%)
Frame = -2
Query: 270 NGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGAEPKKAGAAMASLKDAQRV 329
+G LVT+HGEEAYL+SQLSSFSCIEAGSAEGTAFQGLT+EG EPK+ G AMASLKDAQR
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 330 IQEGQTAGWGKVIQLHENKRKE 351
+QE Q AGWG++IQL ENK K+
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 83.2 bits (204), Expect = 6e-16
Identities = 64/230 (27%), Positives = 108/230 (46%), Gaps = 5/230 (2%)
Frame = +2
Query: 614 VEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKK 673
++ I P P+ R +P +K +++ ++ GF+ YP V ++ + KK
Sbjct: 35 IDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKK 211
Query: 674 DGKVRMCVDFRDLNKASPKDNFPLPHIDMLVDNTAQPKVFSFMDGFSGYNQIKMSPEDRE 733
DG +RM +D+ LN + K +PLP ID L DN K F +D G +Q ++ ED
Sbjct: 212 DGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVP 391
Query: 734 KTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQH 793
KT+F +G + VM FG N + M +F D + V V+ +D+++ S +E +H
Sbjct: 392 KTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEH 571
Query: 794 VEYLTKMFERLRKYKLRLNPNKCTFG-----VRSGKLLGFIVSQKGIEVD 838
+L + L+ L C V G ++S +G++VD
Sbjct: 572 ENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 66.6 bits (161), Expect = 6e-11
Identities = 38/109 (34%), Positives = 55/109 (49%)
Frame = +3
Query: 1101 GAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQI 1160
G + SP + + R+ F +NN YEA I G+ A ++I+++ Y DS LV +Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 1161 KGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSS 1209
GE+E + Y ++L L IPR EN ADALA L+S
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALAS 329
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 59.7 bits (143), Expect = 8e-09
Identities = 33/108 (30%), Positives = 47/108 (42%)
Frame = +2
Query: 1284 RFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWM 1343
R+L D LYK+ D + +RCV E E ++ H + H K+ +AG++W
Sbjct: 17 RYLWDEPFLYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWP 196
Query: 1344 TMEHDCYQHARKCHKCQIYADKIHVPPHALNVISSPWPFSMWGIDMNG 1391
TM D + KC CQ + N I F +WGID G
Sbjct: 197 TMFKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMG 340
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 50.8 bits (120), Expect = 4e-06
Identities = 46/158 (29%), Positives = 71/158 (44%), Gaps = 3/158 (1%)
Frame = +2
Query: 912 PILVPPVEGRPLIMYLVVFDES---MGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTML 968
PILV P LI Y+V D S +GCVL Q E I Y S++ E Y
Sbjct: 17 PILVLP----ELITYVVYTDASITGLGCVLTQH------EKVIAYASRQLRKHEGNYPTH 166
Query: 969 EKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKNARWQMLLSEYDIVFK 1028
+ A+ +A K R YL + + +KYIF + + + RW +++YD+
Sbjct: 167 DLEMAAVVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDI- 343
Query: 1029 TQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLK 1066
T K +++AD L+ + +D E D D + L+
Sbjct: 344 TYYPGKANLVADALSRRRVDVSAEREADDLDGMVRALR 457
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 49.7 bits (117), Expect = 8e-06
Identities = 34/113 (30%), Positives = 51/113 (45%)
Frame = +2
Query: 1086 GLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGIEEAIDMRIK 1145
G V+ +N G GA++++ G + + L T AEY A + G++ A K
Sbjct: 563 G*VW*SNINRGPAGAGALLLAEDGSLLYGFRQGLGHQTKESAEYRALLLGLKHASMKGFK 742
Query: 1146 HLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRDEN 1198
++ GDS LVINQI W+ +L A L F + HI R+ N
Sbjct: 743 YVTAKGDSELVINQILDPWKIKDEHLKKLCAEALELSDNFHSFRIQHISRERN 901
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 48.9 bits (115), Expect = 1e-05
Identities = 24/56 (42%), Positives = 31/56 (54%)
Frame = +1
Query: 931 DESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYL 986
D +G L Q+DE EH I Y S+ T E YT++E+ C A WA + RHYL
Sbjct: 7 DWGIGAGLSQKDEENH-EHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYL 171
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 48.5 bits (114), Expect = 2e-05
Identities = 22/49 (44%), Positives = 30/49 (60%)
Frame = -1
Query: 665 ANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDMLVDNTAQPKVF 713
A ++ V KKDG RMC+D+R NK + K+ +PLP ID L D + F
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF 92
>TC92306
Length = 521
Score = 48.5 bits (114), Expect = 2e-05
Identities = 25/73 (34%), Positives = 37/73 (50%)
Frame = +2
Query: 1133 IFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHH 1192
I G+ EA + +H+ + GD V Q G W+ ++ NL + A L + F V + H
Sbjct: 2 ILGLNEARNQGYEHVHVRGDFQXVCKQFXGSWKVNNPNLRNLCNXAVELKSNFKSVSVEH 181
Query: 1193 IPRDENQMADALA 1205
+PR N ADA A
Sbjct: 182 VPRGXNXAADAQA 220
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides}, partial
(2%)
Length = 780
Score = 36.6 bits (83), Expect = 0.070
Identities = 21/69 (30%), Positives = 32/69 (45%)
Frame = +1
Query: 1139 AIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRDEN 1198
AI + I+ + Y DS L +N IK + HH + + + + LHH R+ N
Sbjct: 496 AISLNIEEMVCYSDSLLTVNLIKEDISQHHVYAVLIHNI--KYIMSSRNFTLHHTLREGN 669
Query: 1199 QMADALATL 1207
Q AD + L
Sbjct: 670 QCADFMVKL 696
>TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~unknown
protein {Arabidopsis thaliana}, partial (69%)
Length = 864
Score = 35.8 bits (81), Expect = 0.12
Identities = 32/111 (28%), Positives = 49/111 (43%), Gaps = 1/111 (0%)
Frame = +2
Query: 164 KSDMLSNVLVDTGSSLNVMPKSTLDQLSYREIPLRRSTFLVKAFDGSRKNV-LGEIDLPI 222
K+D + L D G +++ LD+L R + S F + GSR +G + LPI
Sbjct: 278 KADAAAKELEDAGGLVDLT--DNLDRLQGRWKLIYSSAFSSRTLGGSRPGPPIGRL-LPI 448
Query: 223 TVGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNGKL 273
T+G + D + LG PW VT+TL K + V + K+
Sbjct: 449 TLGQVFQRIDILSKDFDNIVDLQLGAPWPLPPLEVTATLAHKFELVGSSKI 601
>TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG005I10.16 -
Arabidopsis thaliana, partial (19%)
Length = 2073
Score = 34.7 bits (78), Expect = 0.26
Identities = 29/101 (28%), Positives = 49/101 (47%), Gaps = 7/101 (6%)
Frame = -1
Query: 1127 AEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYAR--RLLTY 1184
AE+ AC+ IE+A+++ + ++ + DS V+N H IP++ R + +
Sbjct: 396 AEFCACMIAIEKAMELGLNNICLETDSLKVVNAF------HKIVGIPWQMRVRWHNCIRF 235
Query: 1185 FTKVE--LHHIPRDENQMADALATLS---SMFRVNHWNDVP 1220
+ HIPR+ N +ADALA S+F + W P
Sbjct: 234 CHSIACVCVHIPREGNLVADALARHGQGLSLFFLQWWPAPP 112
>TC84688 similar to GP|10241605|emb|CAC09581. protein kinase {Fagus
sylvatica}, partial (38%)
Length = 1122
Score = 33.1 bits (74), Expect = 0.77
Identities = 27/108 (25%), Positives = 46/108 (42%), Gaps = 6/108 (5%)
Frame = +3
Query: 1264 REYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVL--LRC----VDEHEAEQLMHDV 1317
++Y K D K +R A + L+ + L+ N ++ L+C ++ H+ E + D+
Sbjct: 510 KQYRKKHKKVDLKAVRGWAKQILMGLNYLHTHNPPIIHRDLKCDNIFINGHQGEVKIGDL 689
Query: 1318 HDGTFGTHATGHTMSRKLLRAGYYWMTMEHDCYQHARKCHKCQIYADK 1365
TF A T+ GYYW+T Y A C +D+
Sbjct: 690 GLATFLMQANAKTV------IGYYWITWSFTIYHSAGYKACCSF*SDR 815
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,691,563
Number of Sequences: 36976
Number of extensions: 646601
Number of successful extensions: 3331
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 2875
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3322
length of query: 1391
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1283
effective length of database: 5,021,319
effective search space: 6442352277
effective search space used: 6442352277
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 65 (29.6 bits)
Medicago: description of AC145220.4


