

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145218.2 + phase: 1 /pseudo/partial
(365 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
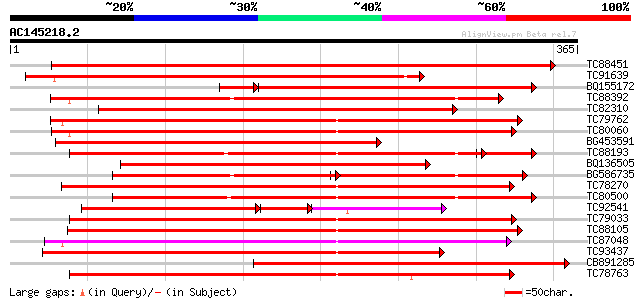
Score E
Sequences producing significant alignments: (bits) Value
TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein ki... 348 1e-96
TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 316 1e-86
BQ155172 weakly similar to GP|6686398|gb F1E22.15 {Arabidopsis t... 296 5e-86
TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-l... 301 3e-82
TC82310 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 291 2e-79
TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.... 290 6e-79
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 282 2e-76
BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-... 278 2e-75
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 257 7e-74
BQ136505 similar to PIR|T05754|T05 S-receptor kinase (EC 2.7.1.-... 265 2e-71
BG586735 similar to GP|11275527|db putative receptor-like protei... 173 1e-69
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 255 2e-68
TC80500 similar to PIR|G96602|G96602 probable receptor protein k... 252 2e-67
TC92541 similar to PIR|T02153|T02153 protein kinase homolog T1F1... 153 2e-67
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 223 7e-59
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 222 2e-58
TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsi... 221 3e-58
TC93437 similar to GP|11227578|emb|CAC16506. unnamed protein pro... 216 8e-57
CB891285 weakly similar to PIR|T14470|T14 receptor-like kinase (... 213 7e-56
TC78763 somatic embryogenesis receptor kinase 1 [Medicago trunca... 211 4e-55
>TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein kinase
{Ipomoea trifida}, partial (28%)
Length = 1276
Score = 348 bits (894), Expect = 1e-96
Identities = 169/324 (52%), Positives = 233/324 (71%)
Frame = +3
Query: 28 ENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQ 87
E+D + ++ + F SI AT FS NKLGQGGYGPVYKG GQEIA+KRLS S Q
Sbjct: 159 EDDFKGHDIKVFNFTSILEATMEFSPENKLGQGGYGPVYKGILATGQEIAVKRLSKTSGQ 338
Query: 88 GLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGW 147
G+ EFKNE++LI +LQH+NLV+L G CI +E+IL+YEYM NKSLD ++FD T+ LL W
Sbjct: 339 GIVEFKNELLLICELQHKNLVQLLGCCIHEEERILIYEYMPNKSLDFYLFDCTKKKLLDW 518
Query: 148 KLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETG 207
K RF+II GIA+G+LYLH+ SRL++IHRDLK SNILLD+ M PKI+DFG+A++F +E+
Sbjct: 519 KKRFNIIEGIAQGLLYLHKYSRLKIIHRDLKASNILLDENMNPKIADFGMARMFTQQESV 698
Query: 208 ASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGY 267
+T R++GTYGYMSPEYA++G S KSDV+SFGV+LLEI+ G KN F+ + +L+G+
Sbjct: 699 VNTNRIVGTYGYMSPEYAMEGVCSTKSDVYSFGVLLLEIVCGIKNNSFYDVDRPLNLIGH 878
Query: 268 AWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETA 327
AW LW + + L LMD L++T +E +C +GLLCV+ +RPTMS ++++L +
Sbjct: 879 AWELWNDGEYLKLMDPTLNDTFVPDEVKRCIHVGLLCVEQYANDRPTMSEVISVLTNKYV 1058
Query: 328 TIPIPSQPTFFTTKHQSCSSSSSK 351
+P +P F+ + ++SK
Sbjct: 1059LTNLPRKPAFYVRREIFEGETTSK 1130
>TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(28%)
Length = 1313
Score = 316 bits (809), Expect = 1e-86
Identities = 155/259 (59%), Positives = 199/259 (75%), Gaps = 2/259 (0%)
Frame = +3
Query: 11 HDSEKHVRDLIGLGNIG--ENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKG 68
H S + ++ G +G N +E+P++ F + +ATNNFS+ NKLGQGG+GPVYKG
Sbjct: 282 HKSREMSAEIPGSVELGLEGNQLSKVELPFFNFSCMSSATNNFSEENKLGQGGFGPVYKG 461
Query: 69 RFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMS 128
+ P G+EIA+KRLS S QGL EFKNE+ L A+LQHRNLV+L G I+GDEK+L+YE+M
Sbjct: 462 KLPSGEEIAVKRLSRRSGQGLDEFKNEMRLFAQLQHRNLVKLMGCSIEGDEKLLVYEFML 641
Query: 129 NKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEM 188
NKSLD F+FD + L W R++II GIARG+LYLH+DSRLR+IHRDLK SNILLD+ M
Sbjct: 642 NKSLDRFLFDPIKKTQLDWARRYEIIEGIARGLLYLHRDSRLRIIHRDLKASNILLDENM 821
Query: 189 IPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILS 248
PKISDFGLA+IFGG + + +V+GTYGYMSPEYA++G S+KSDV+SFGV+LLEI+S
Sbjct: 822 NPKISDFGLARIFGGNQNEENATKVVGTYGYMSPEYAMEGLVSVKSDVYSFGVLLLEIVS 1001
Query: 249 GKKNTGFFRSQQISSLLGY 267
G++NT FR SSL+GY
Sbjct: 1002GRRNTS-FRHSDDSSLIGY 1055
>BQ155172 weakly similar to GP|6686398|gb F1E22.15 {Arabidopsis thaliana},
partial (10%)
Length = 637
Score = 296 bits (758), Expect(2) = 5e-86
Identities = 142/179 (79%), Positives = 162/179 (90%)
Frame = +3
Query: 161 MLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYM 220
+LYLHQDSRLRVIHRDLKTSNILLD+EM PKISDFGLA+I GGKET AST+RV+GTYGYM
Sbjct: 78 LLYLHQDSRLRVIHRDLKTSNILLDEEMQPKISDFGLARIVGGKETEASTERVVGTYGYM 257
Query: 221 SPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDL 280
SPEYALDG+FS KSDVFSFGVVLLEI+SGKKNTGF+RS++ISSLLGYAWRLW E+KL DL
Sbjct: 258 SPEYALDGYFSTKSDVFSFGVVLLEIISGKKNTGFYRSKEISSLLGYAWRLWREDKLQDL 437
Query: 281 MDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPTFFT 339
MD LS+T N N+F++C+QIGLLCVQDEP +RP MSN++TMLD ET T+ P QPTFFT
Sbjct: 438 MDPTLSDTYNVNQFIRCSQIGLLCVQDEPDDRPHMSNVVTMLDNETTTLLTPKQPTFFT 614
Score = 39.7 bits (91), Expect(2) = 5e-86
Identities = 16/25 (64%), Positives = 22/25 (88%)
Frame = +2
Query: 136 IFDRTRTVLLGWKLRFDIIVGIARG 160
IFD T++V+L W +RF+II+GIARG
Sbjct: 2 IFDSTKSVILDWPMRFEIILGIARG 76
>TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(61%)
Length = 1017
Score = 301 bits (771), Expect = 3e-82
Identities = 145/297 (48%), Positives = 215/297 (71%), Gaps = 5/297 (1%)
Frame = +1
Query: 27 GENDSESIEVP-----YYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRL 81
G++D ++++ + ++Q ATN FS+ N+LG+GG+GPV+KG P G+E+AIK+L
Sbjct: 130 GDDDEDTVDPGDSGNLLFELNTLQLATNFFSELNQLGRGGFGPVFKGLMPNGEEVAIKKL 309
Query: 82 SSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTR 141
S S QG++EF NE+ L+ ++QH+NLV L G C +G EK+L+YEY+ NKSLD F+FD+ R
Sbjct: 310 SMESRQGIREFTNEVRLLLRIQHKNLVTLLGCCAEGPEKMLVYEYLPNKSLDHFLFDKKR 489
Query: 142 TVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIF 201
+ L W RF I+ GIARG+LYLH+++ R+IHRD+K SNILLD+++ PKISDFGLA++F
Sbjct: 490 S--LDWMTRFRIVTGIARGLLYLHEEAPERIIHRDIKASNILLDEKLNPKISDFGLARLF 663
Query: 202 GGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQI 261
G++T T R+ T+GYM+PEYAL G+ S+K+DVFS+GV++LEI+SG+KN +
Sbjct: 664 PGEDTHVQTFRISRTHGYMAPEYALRGYLSVKTDVFSYGVLVLEIVSGRKNHDLKLDAEK 843
Query: 262 SSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNI 318
+ LL YAW+L+ K++DL+D + + N +E C Q+GLLC Q RP M+++
Sbjct: 844 ADLLSYAWKLYQGRKIMDLIDQNIGK-YNGDEAAMCIQLGLLCCQASLVERPDMNSV 1011
>TC82310 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(24%)
Length = 696
Score = 291 bits (746), Expect = 2e-79
Identities = 141/231 (61%), Positives = 182/231 (78%)
Frame = +1
Query: 58 GQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKG 117
G+GG+G VYKG +EIA+KRLS S+QG++E KNEI+LIAKLQHRNLVRL CI+
Sbjct: 1 GKGGFGTVYKGVLADEKEIAVKRLSKTSSQGVEELKNEIILIAKLQHRNLVRLLACCIEQ 180
Query: 118 DEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDL 177
+EK+L+YEY+ N SLD +FD + L W+ R +II GIA+G+LYLH+DSRLRVIHRDL
Sbjct: 181 NEKLLIYEYLPNSSLDFHLFDMVKGAQLAWRQRLNIINGIAKGLLYLHEDSRLRVIHRDL 360
Query: 178 KTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVF 237
K SNILLD EM PKISDFGLA+ FGG + A+T RV+GTYGYM+PEYA++G FS+KSDVF
Sbjct: 361 KASNILLDQEMNPKISDFGLARTFGGDQDEANTIRVVGTYGYMAPEYAMEGLFSVKSDVF 540
Query: 238 SFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSET 288
SFGV+LLEI+SG+KN+ F+ S+ SL +AW LW + K +LMD ++ ++
Sbjct: 541 SFGVLLLEIISGRKNSKFYLSEHGQSLPIFAWNLWCKRKGFELMDPSIEKS 693
>TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.8
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1564
Score = 290 bits (742), Expect = 6e-79
Identities = 140/310 (45%), Positives = 219/310 (70%), Gaps = 6/310 (1%)
Frame = +3
Query: 27 GENDSE-----SIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRL 81
G+N++E S E +++ ++ +AT NF+ ++KLG+GG+GPVYKG+ G+E+A+K+L
Sbjct: 297 GDNEAELQKMASREQKIFSYETLLSATKNFNATHKLGEGGFGPVYKGKLSDGREVAVKKL 476
Query: 82 SSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIF-DRT 140
S S QG +EF NE L+A++QH+N+V L GYC+ G EKIL+YEY+ ++SLD F+F +
Sbjct: 477 SQTSNQGKKEFMNEAKLLARVQHKNVVNLLGYCVHGTEKILVYEYVPHESLDKFLFKEAE 656
Query: 141 RTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKI 200
+ L WK RF II G+A+G+LYLH+DS +IHRD+K SNILLDD+ KI+DFG+A++
Sbjct: 657 KREQLDWKRRFGIITGVAKGLLYLHEDSHNCIIHRDIKASNILLDDKWTAKIADFGMARL 836
Query: 201 FGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQ 260
F ++ T RV GT GYM+PEY + G S+K+DVFS+GV++LE+++G++N+ F +
Sbjct: 837 FPEDQSQVKT-RVAGTNGYMAPEYMMHGRLSVKADVFSYGVLVLELITGQRNSSFNLXVE 1013
Query: 261 ISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILT 320
+LL +A++++ + + L+++DSAL+ T + C Q+ LLC+Q +P RPTM I+
Sbjct: 1014EHNLLDWAYKMYKKGRSLEIVDSALASTVLTEQVDMCIQLALLCIQGDPQLRPTMRRIVV 1193
Query: 321 MLDGETATIP 330
L ++ T P
Sbjct: 1194KLSRKSPTKP 1223
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 282 bits (721), Expect = 2e-76
Identities = 145/302 (48%), Positives = 204/302 (67%), Gaps = 3/302 (0%)
Frame = +1
Query: 28 ENDSESIEVP--YYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVS 85
+ D E +E+ YY+ R I+ ATNNF NK+G+GG+GPVYKG G IA+K+LSS S
Sbjct: 118 QTDKELLELKTGYYSLRQIKVATNNFDPKNKIGEGGFGPVYKGVLSDGAVIAVKQLSSKS 297
Query: 86 TQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVL- 144
QG +EF NEI +I+ LQH NLV+L G CI+G++ +L+YEYM N SL +F + L
Sbjct: 298 KQGNREFVNEIGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGKPEQRLN 477
Query: 145 LGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGK 204
L W+ R I VGIARG+ YLH++SRL+++HRD+K +N+LLD + KISDFGLAK+ +
Sbjct: 478 LDWRTRMKICVGIARGLAYLHEESRLKIVHRDIKATNVLLDKNLNAKISDFGLAKLDEEE 657
Query: 205 ETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSL 264
T ST R+ GT GYM+PEYA+ G+ + K+DV+SFGVV LEI+SG NT + ++ L
Sbjct: 658 NTHIST-RIAGTIGYMAPEYAMRGYLTDKADVYSFGVVALEIVSGMSNTNYRPKEEFVYL 834
Query: 265 LGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDG 324
L +A+ L + LL+L+D L + E ++ Q+ LLC P RP MS++++ML+G
Sbjct: 835 LDWAYVLQEQGNLLELVDPTLGSKYSSEEAMRMLQLALLCTNPSPTLRPPMSSVVSMLEG 1014
Query: 325 ET 326
T
Sbjct: 1015NT 1020
>BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-) T6K22.120
precursor - Arabidopsis thaliana, partial (24%)
Length = 640
Score = 278 bits (712), Expect = 2e-75
Identities = 133/211 (63%), Positives = 171/211 (81%), Gaps = 1/211 (0%)
Frame = +3
Query: 30 DSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFP-GGQEIAIKRLSSVSTQG 88
D + E+P++ +I ATN+FS+ NKLG+GG+GPVYKG +EIA+KRLS S QG
Sbjct: 6 DEQDFELPFFNLSTIIDATNDFSNDNKLGEGGFGPVYKGTLVLDRREIAVKRLSGSSKQG 185
Query: 89 LQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWK 148
+EFKNE++L +KLQHRNLV++ G CI+G+EK+L+YEYM N+SLD+F+FD+ + LL W
Sbjct: 186 TREFKNEVILCSKLQHRNLVKVLGCCIQGEEKMLIYEYMPNRSLDSFLFDQAQKKLLDWS 365
Query: 149 LRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGA 208
RF+II GIARG++YLHQDSRLR+IHRDLK SNILLD++M PKISDFGLAKI G +
Sbjct: 366 KRFNIICGIARGLIYLHQDSRLRIIHRDLKPSNILLDNDMNPKISDFGLAKICGDDQVEG 545
Query: 209 STQRVMGTYGYMSPEYALDGFFSIKSDVFSF 239
+T RV+GT+GYM+PEYA+DG FSIKSDVFSF
Sbjct: 546 NTNRVVGTHGYMAPEYAIDGLFSIKSDVFSF 638
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 257 bits (657), Expect(2) = 7e-74
Identities = 129/269 (47%), Positives = 189/269 (69%)
Frame = +3
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVL 98
+++ ++ AT++F+ NKLG+GG+GPVYKG G+ +A+K+LS S QG +F EI
Sbjct: 540 FSYYELKNATSDFNRDNKLGEGGFGPVYKGTLNDGRFVAVKQLSIGSHQGKSQFIAEIAT 719
Query: 99 IAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIA 158
I+ +QHRNLV+L G CI+G++++L+YEY+ NKSLD +F + L W R+D+ +G+A
Sbjct: 720 ISAVQHRNLVKLYGCCIEGNKRLLVYEYLENKSLDQALFG--NVLFLNWSTRYDVCMGVA 893
Query: 159 RGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYG 218
RG+ YLH++SRLR++HRD+K SNILLD E++PK+SDFGLAK++ K+T ST RV GT G
Sbjct: 894 RGLTYLHEESRLRIVHRDVKASNILLDSELVPKLSDFGLAKLYDDKKTHIST-RVAGTIG 1070
Query: 219 YMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLL 278
Y++PEYA+ G + K+DVFSFGVV LE++SG+ N+ + LL +AW+L N +
Sbjct: 1071YLAPEYAMRGRLTEKADVFSFGVVALELVSGRPNSDSSLEEDKMYLLDWAWQLHERNCIN 1250
Query: 279 DLMDSALSETCNENEFVKCAQIGLLCVQD 307
DL+D LSE N E + IG++ D
Sbjct: 1251DLIDPRLSE-FNMEEVERLVGIGIVVHSD 1334
Score = 38.1 bits (87), Expect(2) = 7e-74
Identities = 16/39 (41%), Positives = 24/39 (61%)
Frame = +1
Query: 301 GLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPTFFT 339
GLLC Q P RP+MS ++ ML G+ + S+P ++T
Sbjct: 1315 GLLCTQTSPNLRPSMSRVVAMLLGDIEVSTVTSRPEYWT 1431
>BQ136505 similar to PIR|T05754|T05 S-receptor kinase (EC 2.7.1.-) M4I22.110
precursor - Arabidopsis thaliana, partial (23%)
Length = 618
Score = 265 bits (677), Expect = 2e-71
Identities = 131/200 (65%), Positives = 161/200 (80%)
Frame = +2
Query: 72 GGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKS 131
GG+E+AIKR S +S QGL+EFKNE++LIAKLQHRNLV+L G CI +EK+L+YEYM N+S
Sbjct: 17 GGKEVAIKRNSKMSDQGLEEFKNEVLLIAKLQHRNLVKLLGCCIHREEKLLIYEYMPNRS 196
Query: 132 LDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPK 191
LD FIFD TR+ LL W R II G+ARG+LYLHQDSRLR+IHRDLK SNILLD M PK
Sbjct: 197 LDYFIFDETRSKLLDWSKRSHIIAGVARGLLYLHQDSRLRIIHRDLKLSNILLDALMNPK 376
Query: 192 ISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKK 251
ISDFGLA+ F G + A T++++GTYGYM PEYA+ G +S+KSDVFSFGV++LEI+SGKK
Sbjct: 377 ISDFGLARTFCGDQVEAKTRKLVGTYGYMPPEYAVHGRYSMKSDVFSFGVIVLEIISGKK 556
Query: 252 NTGFFRSQQISSLLGYAWRL 271
F+ +LLG+AWRL
Sbjct: 557 IKVFYDPXHSLNLLGHAWRL 616
>BG586735 similar to GP|11275527|db putative receptor-like protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (57%)
Length = 794
Score = 173 bits (439), Expect(2) = 1e-69
Identities = 81/147 (55%), Positives = 114/147 (77%)
Frame = +3
Query: 67 KGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEY 126
+G P G+E+AIK+LS S QG++EF NE+ L+ ++QH+NLV L G C +G EK+L+YEY
Sbjct: 6 RGLMPNGEEVAIKKLSMESRQGIREFTNEVRLLLRIQHKNLVTLLGCCAEGPEKMLVYEY 185
Query: 127 MSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDD 186
+ NKSLD F+FD+ R+ L W RF I+ GIARG+LYLH+++ R+IHRD+K SNILLD+
Sbjct: 186 LPNKSLDHFLFDKKRS--LDWMTRFRIVTGIARGLLYLHEEAPERIIHRDIKASNILLDE 359
Query: 187 EMIPKISDFGLAKIFGGKETGASTQRV 213
++ PKISDFGLA++F G++T T R+
Sbjct: 360 KLNPKISDFGLARLFPGEDTHVQTFRI 440
Score = 107 bits (268), Expect(2) = 1e-69
Identities = 55/127 (43%), Positives = 81/127 (63%)
Frame = +2
Query: 207 GASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLG 266
G S V GYM+PEYAL G+ S+K+DVFS+GV++LEI+SG+KN + + LL
Sbjct: 413 GHSRANV*DFRGYMAPEYALRGYLSVKTDVFSYGVLVLEIVSGRKNHDLKLDAEKADLLS 592
Query: 267 YAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGET 326
YAW+L+ K++DL+D + + N +E C Q+GLLC Q RP M+++ ML ++
Sbjct: 593 YAWKLYQGGKIMDLIDQNIGK-YNGDEAAMCIQLGLLCCQASLVERPDMNSVNLMLSSDS 769
Query: 327 ATIPIPS 333
T+P PS
Sbjct: 770 FTLPKPS 790
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 255 bits (651), Expect = 2e-68
Identities = 130/293 (44%), Positives = 193/293 (65%), Gaps = 1/293 (0%)
Frame = +3
Query: 34 IEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFK 93
I + YT++ ++ A++NFS +NK+G+GG+G VYKG GG+ AIK LS+ S QG++EF
Sbjct: 258 IRIKVYTYKELKIASDNFSPANKIGEGGFGSVYKGVLKGGKLAAIKVLSTESKQGVKEFL 437
Query: 94 NEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSL-DTFIFDRTRTVLLGWKLRFD 152
EI +I++++H NLV L G C++GD +IL+Y Y+ N SL T + + W+ R
Sbjct: 438 TEINVISEIKHENLVILYGCCVEGDHRILVYNYLENNSLSQTLLAGGHSNIYFDWQTRRR 617
Query: 153 IIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQR 212
I +G+ARG+ +LH++ ++HRD+K SNILLD ++ PKISDFGLAK+ T ST R
Sbjct: 618 ICLGVARGLAFLHEEVLPHIVHRDIKASNILLDKDLTPKISDFGLAKLIPSYMTHVST-R 794
Query: 213 VMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLW 272
V GT GY++PEYA+ G + K+D++SFGV+L+EI+SG+ NT +L W+L+
Sbjct: 795 VAGTIGYLAPEYAIRGQLTRKADIYSFGVLLVEIVSGRSNTNTRLPIADQYILETTWQLY 974
Query: 273 TENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGE 325
+L L+D +L+ + E K +I LLC QD P RPTMS+++ ML GE
Sbjct: 975 ERKELAQLVDISLNGEFDAEEACKILKIALLCTQDTPKLRPTMSSVVKMLTGE 1133
>TC80500 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(13%)
Length = 1015
Score = 252 bits (643), Expect = 2e-67
Identities = 129/273 (47%), Positives = 185/273 (67%)
Frame = +1
Query: 67 KGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEY 126
+G G+++A+K+LS S QG +F EI I+ +QHRNLV+L G CI+G +++L+YEY
Sbjct: 19 QGILNDGRDVAVKQLSIGSHQGKSQFVAEIATISAVQHRNLVKLYGCCIEGSKRLLVYEY 198
Query: 127 MSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDD 186
+ NKSLD +F + L W R+DI +G+ARG+ YLH++SRLR++HRD+K SNILLD
Sbjct: 199 LENKSLDQALFGNV--LFLNWSTRYDICMGVARGLTYLHEESRLRIVHRDVKASNILLDS 372
Query: 187 EMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEI 246
E++PKISDFGLAK++ K+T ST RV GT GY++PEYA+ G + K+DVFSFGVV LE+
Sbjct: 373 ELVPKISDFGLAKLYDDKKTHIST-RVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALEL 549
Query: 247 LSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQ 306
+SG+ N+ + LL +AW+L N + +L+D LSE N+ E + I LLC Q
Sbjct: 550 VSGRPNSDSTLEGEKMYLLEWAWQLHERNTINELIDPRLSE-FNKEEVQRLVGIALLCTQ 726
Query: 307 DEPGNRPTMSNILTMLDGETATIPIPSQPTFFT 339
P RP+MS ++ ML G+ + S+P + T
Sbjct: 727 TSPTLRPSMSRVVAMLSGDIEVGTVTSRPGYLT 825
>TC92541 similar to PIR|T02153|T02153 protein kinase homolog T1F15.1 -
Arabidopsis thaliana, partial (22%)
Length = 791
Score = 153 bits (387), Expect(3) = 2e-67
Identities = 75/115 (65%), Positives = 90/115 (78%)
Frame = +2
Query: 47 ATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRN 106
AT +FS NKLGQGGYGPVYKG P GQEIA+KRLS S QG+ EFKNE+VLI +LQH N
Sbjct: 8 ATIDFSPENKLGQGGYGPVYKGILPTGQEIAVKRLSKTSRQGIVEFKNELVLICELQHTN 187
Query: 107 LVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGM 161
LV+L G CI +E+IL+YEYMSNKSLD ++FD TR L WK R +II GI++G+
Sbjct: 188 LVQLLGCCIHEEERILIYEYMSNKSLDFYLFDSTRRKCLDWKKRLNIIEGISQGL 352
Score = 85.1 bits (209), Expect(3) = 2e-67
Identities = 43/112 (38%), Positives = 65/112 (57%), Gaps = 25/112 (22%)
Frame = +1
Query: 195 FGLAKIFGGKETGASTQRVMGT-------------------------YGYMSPEYALDGF 229
FG+A++F +E+ +T R++GT GYMSPEYA++G
Sbjct: 454 FGMARMFTQQESVVNTNRIVGT**VL*TFLMIT*RKRF*FFK*FFLSSGYMSPEYAMEGI 633
Query: 230 FSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLM 281
S KSDV+SFGV+LLEI+ G++N F+ + +L+G+AW LW + + L LM
Sbjct: 634 CSTKSDVYSFGVLLLEIICGRRNNSFYDVDRPLNLIGHAWELWNDGEYLQLM 789
Score = 56.6 bits (135), Expect(3) = 2e-67
Identities = 26/34 (76%), Positives = 30/34 (87%)
Frame = +3
Query: 162 LYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDF 195
LYLH+ SRL++IHRDLK SNILLD+ M PKISDF
Sbjct: 354 LYLHKYSRLKIIHRDLKASNILLDENMNPKISDF 455
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 223 bits (569), Expect = 7e-59
Identities = 115/289 (39%), Positives = 180/289 (61%), Gaps = 1/289 (0%)
Frame = +1
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVL 98
++ + + +ATNNF+ NKLG+GG+G VY G+ G +IA+KRL S + EF E+ +
Sbjct: 427 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEI 606
Query: 99 IAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRT-VLLGWKLRFDIIVGI 157
+A+++H+NL+ LRGYC +G E++++Y+YM N SL + + + T LL W R +I +G
Sbjct: 607 LARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSTESLLDWNRRMNIAIGS 786
Query: 158 ARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTY 217
A G++YLH + +IHRD+K SN+LLD + +++DFG AK+ T +T RV GT
Sbjct: 787 AEGIVYLHVQATPHIIHRDVKASNVLLDSDFQARVADFGFAKLIPDGATHVTT-RVKGTL 963
Query: 218 GYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKL 277
GY++PEYA+ G + DV+SFG++LLE+ SGKK S ++ +A L E K
Sbjct: 964 GYLAPEYAMLGKANESCDVYSFGILLLELASGKKPLEKLSSSVKRAINDWALPLACEKKF 1143
Query: 278 LDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGET 326
+L D L+ E E + + L+C Q++P RPTM ++ +L GE+
Sbjct: 1144SELADPRLNGDYVEEELKRVILVALICAQNQPEKRPTMVEVVELLKGES 1290
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 222 bits (565), Expect = 2e-58
Identities = 114/294 (38%), Positives = 182/294 (61%), Gaps = 1/294 (0%)
Frame = +2
Query: 38 YYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIV 97
++T R ++ ATN FS N +G+GGYG VY+G+ G +AIK+L + Q +EF+ E+
Sbjct: 686 WFTLRDLELATNKFSKDNIIGEGGYGVVYQGQLINGNPVAIKKLLNNLGQAEKEFRVEVE 865
Query: 98 LIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTV-LLGWKLRFDIIVG 156
I ++H+NLVRL G+CI+G ++L+YEY++N +L+ ++ R L W R I++G
Sbjct: 866 AIGHVRHKNLVRLLGFCIEGTHRLLIYEYVNNGNLEQWLHGAMRQYGYLTWDARIKILLG 1045
Query: 157 IARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGT 216
A+ + YLH+ +V+HRD+K+SNIL+DD+ KISDFGLAK+ G ++ +T RVMGT
Sbjct: 1046TAKALAYLHEAIEPKVVHRDIKSSNILIDDDFNAKISDFGLAKLLGAGKSHITT-RVMGT 1222
Query: 217 YGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENK 276
+GY++PEYA G + KSDV+SFGV+LLE ++G+ + RS +L+ + +
Sbjct: 1223FGYVAPEYANSGLLNEKSDVYSFGVLLLEAITGRDPVDYNRSAAEVNLVDWLKMMVGNRH 1402
Query: 277 LLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIP 330
+++D + + + + L CV + RP MS ++ ML+ E IP
Sbjct: 1403AEEVVDPNIETRPSTSALKRVLLTALRCVDPDSEKRPKMSQVVRMLESEEYPIP 1564
>TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsis
thaliana}, partial (77%)
Length = 1571
Score = 221 bits (564), Expect = 3e-58
Identities = 125/315 (39%), Positives = 189/315 (59%), Gaps = 14/315 (4%)
Frame = +1
Query: 23 LGNIGENDSE-----------SIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFP 71
LGN G N +E S + FR + AT F ++N +G+GG+G V+KGR
Sbjct: 349 LGNGGTNATEEKGKSVRTGKSSTAAASFGFRELATATRGFKEANLIGEGGFGKVFKGRLS 528
Query: 72 GGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKS 131
G+ +A+K+LS QG QEF E+++++ L H NLV+L GYC GD+++L+YEYM S
Sbjct: 529 TGELVAVKQLSHDGRQGFQEFVTEVLMLSLLHHSNLVKLIGYCTDGDQRLLVYEYMPMGS 708
Query: 132 LDTFIFDRTRTVL-LGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIP 190
L+ +FD + L W R I VG ARG+ YLH + VI+RDLK++NILLD + P
Sbjct: 709 LEDHLFDLPQDKEPLSWSSRMKIAVGAARGLEYLHCKADPPVIYRDLKSANILLDSDFSP 888
Query: 191 KISDFGLAKIFG-GKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSG 249
K+SDFGLAK+ G T ST RVMGTYGY +PEYA+ G ++KSD++SFGVVLLE+++G
Sbjct: 889 KLSDFGLAKLGPVGDNTHVST-RVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELITG 1065
Query: 250 KKNTGFFRSQQISSLLGYAWRLWTE-NKLLDLMDSALSETCNENEFVKCAQIGLLCVQDE 308
++ + +L+ ++ +++ K + + D L + I +C+Q++
Sbjct: 1066RRAIDASKKPGEQNLVSWSRPYFSDRRKFVHMADPLLQGHFPVRCLHQAIAITAMCLQEQ 1245
Query: 309 PGNRPTMSNILTMLD 323
P RP + +I+ L+
Sbjct: 1246PKFRPLIGDIVVALE 1290
>TC93437 similar to GP|11227578|emb|CAC16506. unnamed protein product
{Arabidopsis thaliana}, partial (62%)
Length = 978
Score = 216 bits (551), Expect = 8e-57
Identities = 115/261 (44%), Positives = 170/261 (65%), Gaps = 2/261 (0%)
Frame = +3
Query: 22 GLGNIGENDSESI-EVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKR 80
G G E E + + +++ S+++AT +F S K+G GGYG VYKG G ++AIK
Sbjct: 159 GKGEPAEQLHEQVMKTKSFSYNSLRSATGDFHPSCKIGGGGYGVVYKGVLRDGTQVAIKS 338
Query: 81 LSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSL-DTFIFDR 139
LS S QG EF EI +I+ +QH NLV+L G+CI+G+ +IL+YE++ N SL + + +
Sbjct: 339 LSVESKQGTHEFMTEIAMISNIQHPNLVKLIGFCIEGNHRILVYEFLENNSLTSSLLGSK 518
Query: 140 TRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAK 199
++ V L W+ R I G A G+ +LH++++ ++HRD+K SNILLD+ PKI DFGLAK
Sbjct: 519 SKCVPLDWQKRAIICRGTASGLSFLHEEAQPNIVHRDIKASNILLDENFHPKIGDFGLAK 698
Query: 200 IFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQ 259
+F T ST RV GT GY++PEYAL + K+DV+SFG+++LEI+SGK +
Sbjct: 699 LFPDNVTHVST-RVAGTMGYLAPEYALLRQLTKKADVYSFGILMLEIISGKSSXKAAFGD 875
Query: 260 QISSLLGYAWRLWTENKLLDL 280
I L+ +AW+L N+LL+L
Sbjct: 876 NILVLVEWAWKLKEXNRLLEL 938
>CB891285 weakly similar to PIR|T14470|T14 receptor-like kinase (EC 2.7.1.-)
SFR2 - wild cabbage, partial (18%)
Length = 753
Score = 213 bits (543), Expect = 7e-56
Identities = 101/203 (49%), Positives = 145/203 (70%)
Frame = +2
Query: 158 ARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTY 217
ARG+LYLH+DS+LR+IHRDLKTSNILLD+E+ PKISDFG+A+IF G+E +T RV+GTY
Sbjct: 2 ARGLLYLHRDSKLRIIHRDLKTSNILLDEELNPKISDFGMARIFEGREDTENTIRVVGTY 181
Query: 218 GYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKL 277
GY+SPEYA+ G FS KSDVFSFGV+LLEI+SG++N+ F+ ++ +LLG+ W W E +
Sbjct: 182 GYISPEYAMQGLFSEKSDVFSFGVLLLEIISGRRNSSFYDNEHALTLLGFVWIQWKEGNI 361
Query: 278 LDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPTF 337
L +++ + + + V+C IGLLCVQ+ +RP M+ +++ML+ E +P PSQP F
Sbjct: 362 LSFINTEIYDPSHHKYVVRCIHIGLLCVQELAVDRPNMAAVISMLNSEAELLPPPSQPAF 541
Query: 338 FTTKHQSCSSSSSKLEISMQIDS 360
++ + S + I+S
Sbjct: 542 ILRQNMLSTMSHEESHRRYSINS 610
>TC78763 somatic embryogenesis receptor kinase 1 [Medicago truncatula]
Length = 2737
Score = 211 bits (537), Expect = 4e-55
Identities = 118/291 (40%), Positives = 178/291 (60%), Gaps = 4/291 (1%)
Frame = +3
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQ-EFKNEIV 97
++ R +Q AT+ FS+ N LG+GG+G VYKGR G +A+KRL T G + +F+ E+
Sbjct: 1296 FSLRELQVATDTFSNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQTEVE 1475
Query: 98 LIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRT-RTVLLGWKLRFDIIVG 156
+I+ HRNL+RLRG+C+ E++L+Y YM+N S+ + + +R L W R I +G
Sbjct: 1476 MISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPPHQEPLDWPTRKRIALG 1655
Query: 157 IARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGT 216
ARG+ YLH ++IHRD+K +NILLD+E + DFGLAK+ K+T +T V GT
Sbjct: 1656 SARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTT-AVRGT 1832
Query: 217 YGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFR--SQQISSLLGYAWRLWTE 274
G+++PEY G S K+DVF +G++LLE+++G++ R + LL + L E
Sbjct: 1833 IGHIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKE 2012
Query: 275 NKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGE 325
KL L+D L E E + Q+ LLC Q P +RP MS+++ ML+G+
Sbjct: 2013 KKLEMLVDPDLKTNYIEAEVEQLIQVALLCTQGSPMDRPKMSDVVRMLEGD 2165
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,556,901
Number of Sequences: 36976
Number of extensions: 143766
Number of successful extensions: 2045
Number of sequences better than 10.0: 701
Number of HSP's better than 10.0 without gapping: 1434
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1463
length of query: 365
length of database: 9,014,727
effective HSP length: 97
effective length of query: 268
effective length of database: 5,428,055
effective search space: 1454718740
effective search space used: 1454718740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC145218.2


