

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145061.22 - phase: 0 /pseudo
(604 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
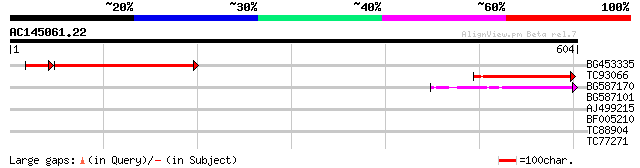
Score E
Sequences producing significant alignments: (bits) Value
BG453335 weakly similar to GP|18071369|g putative gag-pol polypr... 142 1e-40
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 82 8e-16
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 79 5e-15
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 38 0.010
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 30 2.0
BF005210 30 2.7
TC88904 similar to GP|11994550|dbj|BAB02737. gene_id:MGD8.17~unk... 29 5.9
TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-a... 28 7.8
>BG453335 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (11%)
Length = 660
Score = 142 bits (358), Expect(2) = 1e-40
Identities = 86/154 (55%), Positives = 109/154 (69%)
Frame = +2
Query: 48 MHRNAYKELKKKDCKTLFFILLEETL*EFLNPQDQMKLGIFLNGTMKEMLR*SR*SFSY* 107
MH + + ++C + +L++ETL EFLNPQDQ KL IF MK M+R SR*SFS+*
Sbjct: 104 MHIKS*RRRIARNCSSFNRVLMKETLKEFLNPQDQRKLQIFWKNIMKVMIRWSR*SFSH* 283
Query: 108 EGNLS*CKWKMRKRLLITSPC*LML*IR*RFVVEQFLIRK*LRRL**LYLQGLLL*LWPY 167
EG+L+*CKW+MR+RLLITSP * ML*IR*+ + + I K LR+ * YLQ L+L* +
Sbjct: 284 EGSLN*CKWRMRRRLLITSPN*SML*IR*KLMEK*LQINKLLRKS*EHYLQDLIL*WLQF 463
Query: 168 KNQQM*IP*RLKNCIVLWRHMN*WCMKEVVKDQF 201
NQ+M* *RLKN V WR +N*W M+E VKDQF
Sbjct: 464 MNQRM*KL*RLKN*KVPWRLIN*WFMREAVKDQF 565
Score = 42.7 bits (99), Expect(2) = 1e-40
Identities = 20/29 (68%), Positives = 23/29 (78%)
Frame = +3
Query: 18 SVVMKNLFGAKDLFNIVQNGYDELGENPT 46
SVVMKNLF A+DL IVQNGY ++G N T
Sbjct: 3 SVVMKNLFEAQDLLEIVQNGYIDIGANAT 89
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/21 (80%), Positives = 19/21 (89%)
Frame = +1
Query: 47 EMHRNAYKELKKKDCKTLFFI 67
E+HRNA+KELKKKDCK LF I
Sbjct: 91 EVHRNAHKELKKKDCKELFLI 153
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 81.6 bits (200), Expect = 8e-16
Identities = 37/108 (34%), Positives = 67/108 (61%)
Frame = +1
Query: 495 GKQPRTAFISTTTHRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKA 554
G + + +F ST THR+K +L+ ++ D+ GP +V S G +Y ++ +D+F + +WV ++
Sbjct: 43 GNRKKVSF-STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRY 219
Query: 555 KSGAFDIFQKFKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
K+ F F+K++ VE Q+GK++ L TD E+ F +FC +G+
Sbjct: 220 KNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGI 363
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 79.0 bits (193), Expect = 5e-15
Identities = 50/158 (31%), Positives = 83/158 (51%), Gaps = 2/158 (1%)
Frame = -3
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
N+D+ LWH GH + +LN L++P + +K + ++GK + F T+T
Sbjct: 596 NKDA-LWHARLGHPHGRALN--------LMLPGVVFENKNCEACILGKHCKNVFPRTSTV 444
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRI--KYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFK 566
+ +++Y D+ PS+ R KYF++F+DE SK W+ LI +K D F+ F+
Sbjct: 443 Y-ENCFDLIYTDL---WTAPSLSRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQ 276
Query: 567 FFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGVIH 604
+V I LR+D GGEYT F+ + +G++H
Sbjct: 275 AYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILH 162
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 38.1 bits (87), Expect = 0.010
Identities = 32/94 (34%), Positives = 46/94 (48%), Gaps = 1/94 (1%)
Frame = +2
Query: 512 EVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSG-AFDIFQKFKFFVE 570
EV +V D GP PS KY + +D SK WV I + + A + + FK +
Sbjct: 305 EVFDVWGIDFMGPF--PSSYNNKYILVAVDYVSK--WVEAIASPTNDATVVVKMFKSVIF 472
Query: 571 KQSGKSITSLRTDGGGEYTIKVFEKFCEENGVIH 604
+ G + +DGG + KVFEK ++NGV H
Sbjct: 473 PRFGVPRVVI-SDGGSHFINKVFEKLLKKNGVRH 571
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 30.4 bits (67), Expect = 2.0
Identities = 16/48 (33%), Positives = 28/48 (58%)
Frame = +3
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMK 436
+LLSV QL+ KGY + + + D+N + V++ + K + F D+K
Sbjct: 309 SLLSVPQLLTKGYKVLFEHEKCVIKDQNNKEVLRAQM-KARKFVLDLK 449
>BF005210
Length = 650
Score = 30.0 bits (66), Expect = 2.7
Identities = 12/39 (30%), Positives = 23/39 (58%)
Frame = +3
Query: 435 MKVAKLNCLSAIVNNEDSWLWHYMFGHLNYISLNHLVDK 473
M+++ L+ L +V + SW WHY + + N I + ++K
Sbjct: 345 MQLSLLSVLLGVVWGKGSWRWHYFWNYPNMIC*ENTLEK 461
>TC88904 similar to GP|11994550|dbj|BAB02737. gene_id:MGD8.17~unknown protein
{Arabidopsis thaliana}, partial (12%)
Length = 1431
Score = 28.9 bits (63), Expect = 5.9
Identities = 23/83 (27%), Positives = 36/83 (42%), Gaps = 5/83 (6%)
Frame = -1
Query: 436 KVAKLNCLSAIVNNEDSWLWHYMFGHLNYISLNHLVDKEMILVV-----PKLDIPHKFYD 490
KV K CL +I+ +L H+ ++Y+ V + L+V PK+ I + FYD
Sbjct: 1152 KVIKQRCLQSIIFE---YLQHFQTLSIHYLLCVSAVGYRLELIVL*YYVPKISIWYIFYD 982
Query: 491 TRLIGKQPRTAFISTTTHRSKEV 513
L + P + H EV
Sbjct: 981 VPLYREAPTPFILLPEVHILIEV 913
>TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-acid--CoA
ligase (EC 6.2.1.3) isoform 2 - rape, partial (94%)
Length = 2486
Score = 28.5 bits (62), Expect = 7.8
Identities = 19/76 (25%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Frame = -2
Query: 461 HLNYISLNHLVDKEMILVVPKLDIPHKFY-DTRLIGKQPRTAFISTTTHRSKEVLNVVYF 519
H + + + L ++ +V + PH+F D L+G + ++ + H EVL
Sbjct: 1510 HPHLVQEDPLFQAYLVHLVMRQRFPHRFQSDHILVGHEHKSPHVGNFPHARIEVLLQTE* 1331
Query: 520 DVNGPLEVPSMGRIKY 535
DV+ PL + S R ++
Sbjct: 1330 DVHYPLNLASPYRTQF 1283
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.349 0.155 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,547,446
Number of Sequences: 36976
Number of extensions: 232061
Number of successful extensions: 2089
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1192
Number of HSP's successfully gapped in prelim test: 83
Number of HSP's that attempted gapping in prelim test: 883
Number of HSP's gapped (non-prelim): 1308
length of query: 604
length of database: 9,014,727
effective HSP length: 102
effective length of query: 502
effective length of database: 5,243,175
effective search space: 2632073850
effective search space used: 2632073850
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC145061.22


