

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.7 - phase: 0
(482 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
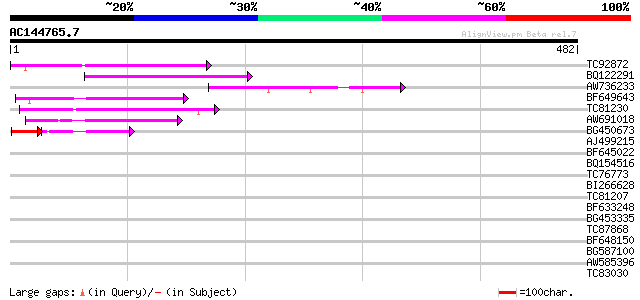
Score E
Sequences producing significant alignments: (bits) Value
TC92872 89 5e-18
BQ122291 88 6e-18
AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Dani... 62 4e-10
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 54 1e-07
TC81230 54 1e-07
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 53 2e-07
BG450673 homologue to GP|12055499|emb NADH dehydrogenase subunit... 39 1e-05
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 40 0.002
BF645022 weakly similar to GP|8777581|dbj retroelement pol polyp... 37 0.022
BQ154516 36 0.029
TC76773 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana ta... 34 0.11
BI266628 34 0.14
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 32 0.41
BF633248 similar to SP|P45598|ARAE_ Arabinose-proton symporter (... 32 0.71
BG453335 weakly similar to GP|18071369|g putative gag-pol polypr... 32 0.71
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 31 0.92
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 31 0.92
BG587100 similar to GP|8777568|dbj contains similarity to copia ... 31 1.2
AW585396 31 1.2
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 30 2.1
>TC92872
Length = 923
Score = 88.6 bits (218), Expect = 5e-18
Identities = 51/173 (29%), Positives = 93/173 (53%), Gaps = 2/173 (1%)
Frame = +1
Query: 1 MASAANNNKNDL--PSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITS 58
MA+ +N K+ L +S++ KL R N+ WK V ++ G ++ G++ GT E I +
Sbjct: 46 MATNPSNTKSSLILNASITFKLSRKNFRAWKRQVTTLLAGIEVMGHIDGTTPIQTETIIN 225
Query: 59 SDSSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRS 118
+ S + NP + +W DQ ++ +L+SM + ET++ LW +S +RS
Sbjct: 226 NGVS-APNPDYTKWFTLDQLIINLLLSSMTEADSISFASYETARTLWVAIESQFNNTSRS 402
Query: 119 QIIYLKSEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGL 171
++ + ++ KG+ + DYL +K+LAD+L + +S+ D+ + LNGL
Sbjct: 403 HVMSVTNQIQRCTKGDKSITDYLFSVKSLADELAVIDKSLSDDDITLFVLNGL 561
>BQ122291
Length = 487
Score = 88.2 bits (217), Expect = 6e-18
Identities = 45/144 (31%), Positives = 84/144 (58%), Gaps = 1/144 (0%)
Frame = -2
Query: 64 SNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYL 123
+ NPA+ EW+ D L W+L+ ++ + ++ + S Q+WDE S +++ L
Sbjct: 465 TRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQMKTRSRQL 286
Query: 124 KSEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLS 183
+SE SI KG + +++ +++ +++ L G+P+S+ DLI L L E++PIV ++
Sbjct: 285 RSELRSITKGSRTVSEFIARIRAISESLASIGDPVSHRDLIEVVLEALPEEFDPIVASVN 106
Query: 184 DHT-TLSWVDLQAQLLTFESRIEQ 206
+ +S +L++QLLT ESR E+
Sbjct: 105 AKSEVVSLDELESQLLTQESRKEK 34
>AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Danio rerio},
partial (14%)
Length = 510
Score = 62.4 bits (150), Expect = 4e-10
Identities = 50/179 (27%), Positives = 85/179 (46%), Gaps = 12/179 (6%)
Frame = +1
Query: 170 GLDSEYNPIVVKLSDHT-TLSWVDLQAQLLTFESRIEQLNN-LTNLNLNAT---ANVANK 224
GL EYN V+ + + S + ++ L+ E++ E+ LTN +++A + + +K
Sbjct: 1 GLLEEYNSFVLVIYIRLDSPSMEEGESILMMQEAQFEKYRQELTNPSVSANLAQSELPSK 180
Query: 225 FDHRDNRFNSNNNWRGSNFRGWRGGRGRGR----SSKAPCQVCGKTNHTAINCFHRFDKN 280
+ +++ + RG GRGRGR S++ CQ+C + NH A C R+D+
Sbjct: 181 PGNSESQEVGTEYYNAGRGRGRGRGRGRGRGRSNSNRLQCQICARNNHDAARCCFRYDQ- 357
Query: 281 YSRSNYSADSDKQGSHNA---FIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHG 336
S Q H A A+ +S + WY DSGAS+H+T + T ++G
Sbjct: 358 --------ASSSQAHHRAPPSEYAASSSYSEAPWYPDSGASHHLTFNPHNLAYRTPYNG 510
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 54.3 bits (129), Expect = 1e-07
Identities = 36/163 (22%), Positives = 70/163 (42%), Gaps = 16/163 (9%)
Frame = +1
Query: 6 NNNKNDLPSS---------------VSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKK 50
N N +D+ SS V KL+ NY W ++ + G++ G+ K
Sbjct: 88 NKNNSDMDSSNPYYIHPSDHPGHLLVPTKLNGTNYSSWSRSMVHALTAKNKVGFINGSIK 267
Query: 51 CPEEFITSSDSSKSNNPA-FEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQ 109
P E + PA + W + +L W+ +S+ ++A ++H +T+ Q+W++ +
Sbjct: 268 TPSEV---------DQPAEYALWNRCNSMILSWLTHSVEPDLAKGVIHAKTACQVWEDFK 420
Query: 110 SLAGAHTRSQIIYLKSEFHSIRKGEMKMEDYLIKMKNLADKLK 152
I ++ S+ +G + Y K+K L D+L+
Sbjct: 421 DQFSQKNIPAIYQIQKSLASLSQGTVSASTYFTKIKGLWDELE 549
>TC81230
Length = 958
Score = 53.9 bits (128), Expect = 1e-07
Identities = 40/183 (21%), Positives = 80/183 (42%), Gaps = 13/183 (7%)
Frame = +1
Query: 9 KNDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPA 68
K ++ +S+ L+ +NY W + ++G +L Y+ G KKCP + D++ +
Sbjct: 205 KLEVTPPISIILNGSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTK--GKDDTADAFADK 378
Query: 69 FEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDE-AQSLAGAHTRSQIIYLKSEF 127
EEW + + +++ W N+ + Q E +K++WD Q + Q LK
Sbjct: 379 LEEWDSKNHQIITWFRNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQYQLLKDLS 558
Query: 128 HSIRKGEMKMEDYLIKMKNLADKLKLAGNPIS------------NSDLIIQTLNGLDSEY 175
+ ++ + ++L +M+ + ++L + N +IQ L L EY
Sbjct: 559 NLKQQSGQPVYEFLAQMEVIWNQLTSCEPSLKDATDMKTYETHRNRVRLIQFLMALTDEY 738
Query: 176 NPI 178
P+
Sbjct: 739 EPV 747
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 53.1 bits (126), Expect = 2e-07
Identities = 34/135 (25%), Positives = 66/135 (48%), Gaps = 1/135 (0%)
Frame = +2
Query: 14 SSVSVKLDRNNYPLW-KSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEW 72
S V+ L+ NY W +S++L + KL G + GT K P +++P W
Sbjct: 191 SLVNQPLNGGNYGEWSRSMLLSLSAKNKL-GLIDGTVKAPS----------ADDPKLPLW 337
Query: 73 QANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRK 132
+ + +L W+L+S+ ++A ++ +T+ +W + S+I ++ E R+
Sbjct: 338 KRCNDLVLTWILHSIEPDIARSVIFSDTAAAVWSDLHDRFSQGDESRIYQIRQEISECRQ 517
Query: 133 GEMKMEDYLIKMKNL 147
G + + DY K+K+L
Sbjct: 518 GSLLISDYYTKLKSL 562
>BG450673 homologue to GP|12055499|emb NADH dehydrogenase subunit 2 {Sphagnum
fallax}, partial (2%)
Length = 678
Score = 38.9 bits (89), Expect(2) = 1e-05
Identities = 21/79 (26%), Positives = 37/79 (46%)
Frame = +3
Query: 28 WKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQRLLGWMLNSM 87
W+ L P+V G GY+ G P + +P F W ++ + GW++NSM
Sbjct: 201 WQQLS-PLVSGKDKLGYINGDLPPPPQ----------TDPTFRRWGTDNAIVKGWIINSM 347
Query: 88 ATEMATQLLHCETSKQLWD 106
+ + + + T+K +WD
Sbjct: 348 DSSLISNFIRFSTTKLVWD 404
Score = 28.1 bits (61), Expect(2) = 1e-05
Identities = 10/27 (37%), Positives = 17/27 (62%)
Frame = +1
Query: 2 ASAANNNKNDLPSSVSVKLDRNNYPLW 28
A ++N + + + +KLD +NYPLW
Sbjct: 142 APPPHSNSDHTVAPIGIKLDGSNYPLW 222
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 40.4 bits (93), Expect = 0.002
Identities = 23/76 (30%), Positives = 42/76 (55%), Gaps = 4/76 (5%)
Frame = +3
Query: 311 WYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKS----LNLDDV 366
W DSG ++H+TH + F++L + ++ NG +E+ + +KS + +V
Sbjct: 108 WLIDSGCTHHMTHDRDLFKELNKSTISKVRML-NGAHIEVEGIGTVLVKSHSGYKQISNV 284
Query: 367 LYVPNITKNLLSVSKL 382
LY P + ++LLSV +L
Sbjct: 285 LYAPKLNQSLLSVPQL 332
>BF645022 weakly similar to GP|8777581|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (8%)
Length = 685
Score = 36.6 bits (83), Expect = 0.022
Identities = 26/127 (20%), Positives = 55/127 (42%), Gaps = 2/127 (1%)
Frame = -2
Query: 10 NDLPSSV--SVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNP 67
ND P +V ++L NY W + + + G++ G K P+ +
Sbjct: 360 NDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEG--KIPKP---------TTPE 214
Query: 68 AFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEF 127
E+W+A L+ W+LN++ + + L + + ++ LW + ++I LK+
Sbjct: 213 KLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASL 34
Query: 128 HSIRKGE 134
++G+
Sbjct: 33 GECKQGK 13
>BQ154516
Length = 506
Score = 36.2 bits (82), Expect = 0.029
Identities = 15/45 (33%), Positives = 27/45 (59%)
Frame = +3
Query: 301 ASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNG 345
A+ +S + WY DSGAS+H+T + T ++G+ +++ NG
Sbjct: 96 AASSSYSEAPWYPDSGASHHLTFNPHNLAYRTPYNGQEQVLMDNG 230
>TC76773 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana tabacum},
partial (86%)
Length = 1228
Score = 34.3 bits (77), Expect = 0.11
Identities = 38/137 (27%), Positives = 54/137 (38%), Gaps = 3/137 (2%)
Frame = +2
Query: 3 SAANNNKNDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITS---S 59
S+A N N+L S L +N PL + V + K +F TS
Sbjct: 149 SSAARNLNELVQDQSQLLHYHNGPLLYGKIS--VNLIWYGNFKPSQKAIITDFFTSLSSP 322
Query: 60 DSSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQ 119
SSK N P+ W ++ +T+++ L +KQ+ DE SL + T
Sbjct: 323 SSSKPNQPSVSAWWKTTEKYYHLTSKKKSTQLSLSL-----NKQILDENYSLGKSLTNKN 487
Query: 120 IIYLKSEFHSIRKGEMK 136
II L S KGE K
Sbjct: 488 IIQLAS------KGEHK 520
>BI266628
Length = 536
Score = 33.9 bits (76), Expect = 0.14
Identities = 24/115 (20%), Positives = 52/115 (44%), Gaps = 7/115 (6%)
Frame = -1
Query: 16 VSVKLDRNNYPL-------WKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPA 68
V +K+D +N + WK V V+R K +++ + P + + + PA
Sbjct: 455 VVIKVDPSNKQVLS*VRLQWKQQVEGVLRETKTVKFVISPQIPPVYLTDEAREAGTKKPA 276
Query: 69 FEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYL 123
+ EW+ D L W+L+++ + ++ + S W + + A +++I+ L
Sbjct: 275 YTEWEEQDSLLCTWILSTI*PSLLSRFVLLRHS---WFGMRFIVTASCKAKILAL 120
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 32.3 bits (72), Expect = 0.41
Identities = 18/46 (39%), Positives = 24/46 (52%)
Frame = +3
Query: 235 NNNWRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCFHRFDKN 280
+N+ G RG+RGG R + C CG T H A +C R D+N
Sbjct: 282 DNHGGGGGGRGFRGGERR--NGGGGCYTCGDTGHIARDC-DRSDRN 410
>BF633248 similar to SP|P45598|ARAE_ Arabinose-proton symporter (Arabinose
transporter). {Klebsiella oxytoca}, partial (5%)
Length = 514
Score = 31.6 bits (70), Expect = 0.71
Identities = 14/42 (33%), Positives = 24/42 (56%)
Frame = +1
Query: 60 DSSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETS 101
DSSK N+ E+W ++ R++ M+N+ AT T ++ S
Sbjct: 388 DSSKKNSHGSEKWISSKMRVMNKMINTTATVATTPIMRPNNS 513
>BG453335 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (11%)
Length = 660
Score = 31.6 bits (70), Expect = 0.71
Identities = 17/67 (25%), Positives = 37/67 (54%), Gaps = 3/67 (4%)
Frame = +1
Query: 134 EMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKL---SDHTTLSW 190
E K+ DY+ K+ N+ +++K G +++ ++ + + L ++ IVV + D TL
Sbjct: 316 EKKIADYISKLINVVNQIKAYGEVVTDQQIVEKIMRTLSPRFDFIVVAIHESKDVKTLKI 495
Query: 191 VDLQAQL 197
+L++ L
Sbjct: 496 EELKSSL 516
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 31.2 bits (69), Expect = 0.92
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 4/47 (8%)
Frame = +3
Query: 234 SNNNWRGSNFRGWRGGRGR----GRSSKAPCQVCGKTNHTAINCFHR 276
S+N+ G G GGRGR G S C CG+ H A C +R
Sbjct: 246 SHNSRSGGGGGGGGGGRGRGGGGGGGSDLKCYECGEPGHFARECRNR 386
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 31.2 bits (69), Expect = 0.92
Identities = 32/142 (22%), Positives = 60/142 (41%), Gaps = 2/142 (1%)
Frame = +2
Query: 70 EEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHS 129
++W +D LG +LN M+ + ++K LWD+ ++ + +L S F++
Sbjct: 173 QKWDNDDYICLGHILNGMSDSLFDIYQSSPSAKDLWDKLETRYMREDATSKKFLVSHFNN 352
Query: 130 IRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHTTLS 189
KM D M+ L + ++ N ++ + +T+ IV + D S
Sbjct: 353 -----YKMVDNKSVMEQLYEIERILNNYKQHNMNMDETI---------IVSSIIDKLPPS 490
Query: 190 WVDLQAQLLTFESRI--EQLNN 209
W D + + + I EQL N
Sbjct: 491 WKDFKRTMKHKKEDISLEQLGN 556
>BG587100 similar to GP|8777568|dbj contains similarity to copia
polyprotein~gene_id:T13B17.2 {Arabidopsis thaliana},
partial (51%)
Length = 782
Score = 30.8 bits (68), Expect = 1.2
Identities = 30/141 (21%), Positives = 61/141 (42%), Gaps = 1/141 (0%)
Frame = +3
Query: 75 NDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGE 134
N+ + + + + +A + Q L E +LW E ++ + ++ ++R +
Sbjct: 246 NNYKAIVVIRHHLAEGLKDQYLTIENPLELWTELKTRFNHQKTVILPKALYDWRNLRIQD 425
Query: 135 MK-MEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDL 193
K +E+Y + + KLKL G I+++D++ +T S ++ V L
Sbjct: 426 YKSVEEYNSALFKIVSKLKLCGETITDADMLEKTF----STFHTSNVVLQQQYREKGFST 593
Query: 194 QAQLLTFESRIEQLNNLTNLN 214
A L++ EQ N L +N
Sbjct: 594 YAALISCLLLAEQNNELLIMN 656
>AW585396
Length = 555
Score = 30.8 bits (68), Expect = 1.2
Identities = 26/83 (31%), Positives = 43/83 (51%), Gaps = 1/83 (1%)
Frame = +1
Query: 145 KNLADKLKLAGNP-ISNSDLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDLQAQLLTFESR 203
+NL + +K N I+ +I +T N LDS V+ + H T S Q+ L + +
Sbjct: 88 ENLQNSIKSISNENIAFKAIIEETRNSLDSS-----VEENAHLTTSLEKSQSLL---KLK 243
Query: 204 IEQLNNLTNLNLNATANVANKFD 226
++LNNLTNL N+ +++ D
Sbjct: 244 TKELNNLTNLMENSKRSLSKNSD 312
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 30.0 bits (66), Expect = 2.1
Identities = 18/53 (33%), Positives = 24/53 (44%), Gaps = 11/53 (20%)
Frame = +1
Query: 232 FNSNNNW----RGSNFRGWRGGRGRGRSSKAP-------CQVCGKTNHTAINC 273
+ S N+W R S GGR RSS +P C CG++ H A +C
Sbjct: 49 YKSGNSWSKPERSSRDDWLIGGRQSSRSSSSPNRSFAGTCFTCGESGHRASDC 207
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.130 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,557,658
Number of Sequences: 36976
Number of extensions: 195054
Number of successful extensions: 1234
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 1213
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1227
length of query: 482
length of database: 9,014,727
effective HSP length: 100
effective length of query: 382
effective length of database: 5,317,127
effective search space: 2031142514
effective search space used: 2031142514
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144765.7


