

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
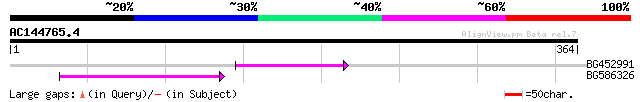
Query= AC144765.4 - phase: 0 /pseudo
(364 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 45 3e-05
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 42 3e-04
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 45.4 bits (106), Expect = 3e-05
Identities = 35/72 (48%), Positives = 39/72 (53%)
Frame = +1
Query: 146 LNSLEI*A*FASGHLRV*N*EC*RLTVNF*RVLKRHRKLMSSL*TCWLLVIRLKMVTLKS 205
LNS E * F RV*N C*R T NF V KR R+ M S * L +IRLKMV LK
Sbjct: 70 LNSSET*VWFVKCRHRV*NWGC*RSTTNFWIVSKRLRRWM*SW*I*CLGIIRLKMVILKL 249
Query: 206 MTMVC*DSEVEF 217
M C +E+
Sbjct: 250 MIKECCSFGIEY 285
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 42.4 bits (98), Expect = 3e-04
Identities = 42/106 (39%), Positives = 50/106 (46%)
Frame = +3
Query: 33 LWFTVMRPSWV*EVYLCKMVKW*LMLQGS*ELMRRITRRMI*SWRLWSLY*RYGDITCMV 92
+WF R S * Y M + QGS*E MR T MI* W *R+G TCMV
Sbjct: 48 MWFIQTRLSLD*VAY*PSMRRSSPTRQGS*ENMRETTPPMI*KWLR*YSP*RFGAHTCMV 227
Query: 93 RGLRCLVITRV*SICSIRKN*I*GNEGGSNY*KIMTLI*IIIQVKL 138
R + +V*SI +* *G GG N * +II+ KL
Sbjct: 228 PRFRYIRTIKV*SIFLPSLS*T*GRGGGWNS*LTTI*TSLIIREKL 365
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.379 0.173 0.672
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,892,006
Number of Sequences: 36976
Number of extensions: 151762
Number of successful extensions: 1786
Number of sequences better than 10.0: 4
Number of HSP's better than 10.0 without gapping: 671
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 1107
Number of HSP's gapped (non-prelim): 745
length of query: 364
length of database: 9,014,727
effective HSP length: 97
effective length of query: 267
effective length of database: 5,428,055
effective search space: 1449290685
effective search space used: 1449290685
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 13 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 35 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144765.4


